Genetic Prediction of Male Pattern Baldness
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Groom Your Man!
GROOM YOUR MAN! When it comes to grooming, women have it down to a science. But what about the man in your life? Kyan Douglas, the grooming guru for "Queer Eye for the Straight Guy," stopped by The Early Show on Wednesday, April 8, 2009, to discuss how to have THE conversation with your man about grooming issues. He then addressed the key grooming tools every man should have!: DO YOU THINK MOST MEN ARE EVEN CONCERNED OR REALIZE THEY NEED TO TAKE CARE OF THESE GROOMING ISSUES? With women, they have a sense of their appearance, some guys are all over it....very metro men are all about products.....but we're in the middle of a transition of what is appropriate for men and they're grooming habits ...men do care, but it's still a mixed bag of how far they take it with products and grooming. I always tell men that they don't want to ever look like they are neglectful of their appearance. IS THERE EVER A GOOD TIME TO BROACH THE SUBJECT OF GROOMING WITH YOUR MAN? First of all, when you're in a relationship you have to remind yourself and your partner this is primarily coming from a place of loving your mate. No matter what, he has to feel there has to be unconditional acceptance and there has to be a tone of safety in your relationship before you can talk about something like this. HOW DO YOU TELL YOUR MAN THAT HE REALLY NEEDS TO DO SOMETHING ABOUT HIS 1.NOSE HAIR, 2. -

PLATFORM ABSTRACTS Abstract Abstract Numbers Numbers Tuesday, November 6 41
American Society of Human Genetics 62nd Annual Meeting November 6–10, 2012 San Francisco, California PLATFORM ABSTRACTS Abstract Abstract Numbers Numbers Tuesday, November 6 41. Genes Underlying Neurological Disease Room 134 #196–#204 2. 4:30–6:30pm: Plenary Abstract 42. Cancer Genetics III: Common Presentations Hall D #1–#6 Variants Ballroom 104 #205–#213 43. Genetics of Craniofacial and Wednesday, November 7 Musculoskeletal Disorders Room 124 #214–#222 10:30am–12:45 pm: Concurrent Platform Session A (11–19): 44. Tools for Phenotype Analysis Room 132 #223–#231 11. Genetics of Autism Spectrum 45. Therapy of Genetic Disorders Room 130 #232–#240 Disorders Hall D #7–#15 46. Pharmacogenetics: From Discovery 12. New Methods for Big Data Ballroom 103 #16–#24 to Implementation Room 123 #241–#249 13. Cancer Genetics I: Rare Variants Room 135 #25–#33 14. Quantitation and Measurement of Friday, November 9 Regulatory Oversight by the Cell Room 134 #34–#42 8:00am–10:15am: Concurrent Platform Session D (47–55): 15. New Loci for Obesity, Diabetes, and 47. Structural and Regulatory Genomic Related Traits Ballroom 104 #43–#51 Variation Hall D #250–#258 16. Neuromuscular Disease and 48. Neuropsychiatric Disorders Ballroom 103 #259–#267 Deafness Room 124 #52–#60 49. Common Variants, Rare Variants, 17. Chromosomes and Disease Room 132 #61–#69 and Everything in-Between Room 135 #268–#276 18. Prenatal and Perinatal Genetics Room 130 #70–#78 50. Population Genetics Genome-Wide Room 134 #277–#285 19. Vascular and Congenital Heart 51. Endless Forms Most Beautiful: Disease Room 123 #79–#87 Variant Discovery in Genomic Data Ballroom 104 #286–#294 52. -

Facial Image Comparison Feature List for Morphological Analysis
Disclaimer: As a condition to the use of this document and the information contained herein, the Facial Identification Scientific Working Group (FISWG) requests notification by e-mail before or contemporaneously to the introduction of this document, or any portion thereof, as a marked exhibit offered for or moved into evidence in any judicial, administrative, legislative, or adjudicatory hearing or other proceeding (including discovery proceedings) in the United States or any foreign country. Such notification shall include: 1) the formal name of the proceeding, including docket number or similar identifier; 2) the name and location of the body conducting the hearing or proceeding; and 3) the name, mailing address (if available) and contact information of the party offering or moving the document into evidence. Subsequent to the use of this document in a formal proceeding, it is requested that FISWG be notified as to its use and the outcome of the proceeding. Notifications should be sent to: Redistribution Policy: FISWG grants permission for redistribution and use of all publicly posted documents created by FISWG, provided the following conditions are met: Redistributions of documents, or parts of documents, must retain the FISWG cover page containing the disclaimer. Neither the name of FISWG, nor the names of its contributors, may be used to endorse or promote products derived from its documents. Any reference or quote from a FISWG document must include the version number (or creation date) of the document and mention if the document is in a draft status. Version 2.0 2018.09.11 Facial Image Comparison Feature List for Morphological Analysis 1. -

Hypertrichosis in a Patient with Hemophagocytic Lymphohistiocytosis Jeylan El Mansoury A, Joyce N
+ MODEL International Journal of Pediatrics and Adolescent Medicine (2015) xx,1e2 HOSTED BY Available online at www.sciencedirect.com ScienceDirect journal homepage: http://www.elsevier.com/locate/ijpam WHAT’S YOUR DIAGNOSIS Hypertrichosis in a patient with hemophagocytic lymphohistiocytosis Jeylan El Mansoury a, Joyce N. Mbekeani b,c,* a Dept of Ophthalmology, KFSH&RC, Riyadh, Saudi Arabia b Dept of Surgery, Jacobi Medical Center, North Bronx Health Network, Bronx, NY, USA c Dept of Ophthalmology and Visual Sciences, Albert Einstein College of Medicine of Yeshiva University, Bronx, NY, USA Received 3 March 2015; received in revised form 15 April 2015; accepted 1 May 2015 KEYWORDS Cyclosporine A; Hypertrichosis; Trichomegaly; Hemophagocytic lymphohistiocytosis; Iatrogenic 1. A case of pediatric hypertrichosis and the diagnosis of HLH. Molecular genetic testing was nega- trichomegaly tive thus ruling out hereditary HLH. Intravenous cyclo- sporine A (CSA) and prednisolone were administered per HLH-2004 protocol for 2 weeks. He was discharged in stable A 19 month old baby boy was referred to ophthalmology for condition on oral CSA 100 mg BID in preparation for sub- pre-operative assessment prior to bone marrow transplant sequent allogeneic BMT. Ophthalmology examination (BMT) for hemophagocytic lymphohistiocytosis (HLH). This revealed an alert, healthy looking baby boy with eyes that rare disorder of deregulated cellular immunity was diag- were central, steady, maintained and could fix and follow nosed at 8 months when he presented with failure to small objects; the anterior and posterior segments were thrive, hepatosplenomegaly, pancytopenia and elevated normal. Abnormally long and thick crown of head hair, liver function tests. Bone marrow biopsy revealed multiple eyelashes (or trichomegaly) and a thick unibrow were histiocytes with hemophagocytic activity compatible with noted (Fig. -
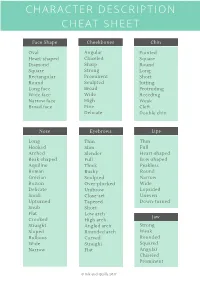
Character Description Cheat Sheet
CHARACTER DESCRIPTION CHEAT SHEET Face Shape Cheekbones Chin Oval Angular Pointed Heart-shaped Chiseled Square Diamond Sharp Round Square Strong Long Rectangular Prominent Short Round Sculpted Jutting Long face Broad Protruding Wide face Wide Receding Narrow face High Weak Broad face Fine Cleft Delicate Double chin Nose Eyebrows Lips Long Thin Thin Hooked Slim Full Arched Slender Heart-shaped Beak-shaped Full Bow-shaped Aquiline Thick Peakless Roman Bushy Face Shape RoundF ace Shape Grecian Sculpted Narrow Button Over-plucked Wide Delicate Unibrow Lopsided Small Close-set Uneven Upturned Tapered Down-turned Snub Short Flat Low arch Jaw Crooked High arch Straight Angled arch Strong Sloped Rounded arch Weak Bulbous Curved Rounded Wide Straight Squared Narrow Flat Angular Chiseled Prominent © Ink and Quills 2017 CHARACTER DESCRIPTION CHEAT SHEET Eye Shape Body Skin Tone Almond Thin Porcelain Round Slim Alabaster Deep-set Slender Ivory Monolid Skinny Fair Hooded Scrawny Pale eyelids Lean Pallid Droopy Wiry Ruddy eyelids Petite Rosy Downturned Dainty Beige Upturned Willowy Tawny Protruding Lanky Umber Close set Gangling Brown Wide set Curvy Sepia Full-figured Sienna Athletic Bronze Eye Color Muscular Topaz Sculpted Amber Amber Toned Tanned Hazel Strong Golden Chestnut Brawny Copper Chocolate Hulking Ocher Honey-brown Beefy Dusky Topaz Barrel-chested Swarthy Russet Stocky Ebony Emerald green Squat Obsidian Forest green Stout Midnight Sea green Burly Olive green Portly Jade Pudgy Blue-green Skin Misc. Overweight Sapphire Husky Tattoos Cornflower -

Dermalaser Digital Brochure
" Safety and Efficacy of our treatments are the cornerstones upon which this dedicated laser clinic is built " D r M i c h a e l D a v i s • D r I a n W e b s t e r • D r C o r d u l a W i n k l e r • D r P i e r r e v a n Z y l Dermalaser Medical Skin and Laser Clinic | Somerset West Understanding who we are Dermalaser Medical Skin & Laser Clinic is a leading provider of specialist skin treatments based in the picturesque town of Somerset West in the Cape Winelands. The clinic is owned by Plastic & Reconstructive Surgeon; Dr Winkler and three Dermatologists; Dr Mike Davis, Dr Ian Webster and Dr Pierre van Zyl who collectively bring 40 years worth of laser experience and expertise to the clinic. Keeping up with state-of-the-art latest technology, the clinic owns some of the best medical lasers available. Our skin and aesthetic laser therapists work under the guidance and strict protocols of our four medical specialists. As registered members of the South African Dermatology Physician Assistant‘s Association, all staff receive ongoing, innovative, evidence-based educational training by specialists in the field of Dermatology, enabling us to provide the highest quality professional patient care. Extensive training and years of experience, combined with great communication skills makes Dermalaser Medical Skin & Laser Clinic the clinic of choice for discerning clients. Dermalaser Medical Skin and Laser Clinic | About Us W h a t e v e r y o u r W e h a v e a Skin Concerns Treatment Acne................................................................. -

Eyelash-Enhancing Products: a Review Bishr Al Dabagh, MD; Julie Woodward, MD
Review Eyelash-Enhancing Products: A Review Bishr Al Dabagh, MD; Julie Woodward, MD Long prominent eyelashes draw attention to the eyes and are considered a sign of beauty. Women strive to achieve ideal lashes through various modalities, including cosmetics, cosmeceuticals, and pharmaceu- ticals. The booming beauty industry has introduced many cosmetic and cosmeceutical products with claims of enhancing eyelash growth; however, many of these products have not been tested for efficacy or safety and are promoted solely with company- and/or consumer-based claims. The only pharmaceu- tical approved by the US Food and Drug Administration for eyelash growth is bimatoprost ophthalmic solution 0.03% (Latisse, Allergan, Inc). In this article, eyelash physiology, causes of genetic and acquired trichomegaly, and pharmaceutical and cosmeceutical products that claim eyelash-enhancing effects are reviewed. COS DERMCosmet Dermatol. 2012;25:134-143. yelashes decorate the eyes and crystallize cosmeceuticals contain active ingredients that are intended the beauty of the face.1 Long and lush eye- to produce beneficial physiologic effects through medicinal Dolashes in particular Notare considered a sign properties. Copy3,4 Cosmeceuticals fall in the gray area between of beauty. Women strive to achieve more inert cosmetics and pharmaceuticals. Drugs are defined pronounced eyelashes using a variety of tech- by the FDA as “articles intended for use in the diagnosis, niques.E Cosmetics are the mainstay of eyelash enhance- cure, mitigation, treatment, or prevention -

How Does the Representation of Women in Billie's Advertisement
How does the representation of women in Billie’s advertisement affect its message of empowerment? An analysis of the campaign video, “Project Body Hair” 11. December 2018 Roskilde Universitet Claudia Maria Bednorz - 64633 Rebecca Elise Glitfeldt - 35492 Dario Napoli - 64284 Christina Juhl Pausgaard - 64914 Elsa Lea Cecile Perrin - 62524 Group Project in Communication Studies, Autumn 2018 Page 1 of 71 Total Characters: 140355 Table of Contents I. Introduction .......................................................................................................................... 4 A. Research question and approach ...................................................................................... 6 B. Relevance ......................................................................................................................... 7 C. Structure ........................................................................................................................... 8 II. Methodological Approaches ................................................................................................ 8 A. Feminist Research Ethic ................................................................................................... 9 1. Epistemology .................................................................................................................. 11 2. Boundaries and Limitations ........................................................................................... 12 3. Relationships ................................................................................................................. -
Address Get in Touch Hours Follow Us
HAIRCUT & STYLE ADDRESS Women Men 3221 Derry Rd. W, Unit 19, Bang Trim $13 Regular Cut $25 Straight Cut $24 Zero Fade Cut $35 Mississauga, Ontario U-Shape & V-Shape Cut $30 Stylist Cut $35+ Front Layer Cut $35 Beard Trim $15 L5N 7L7 All Over Layers Cut $40 Beard Line $20 Bob Cut $40+ Haircut & Colour $55 Stylist Cut $50+ Haircut & Ammonia-Free Colour $65 Short Hair Curls & Straight Blow Dry $30+ Highlights $85 Medium Hair Curls & Straight Blow Dry $45+ HOURS Long Hair Curls & Straight Blow Dry $55+ Shampoo $10 Kids (Ages 0 - 12) For salon hours, Wash & Fluff Dry $20 please visit bellussalon.ca Head Oil Massage (15 Min) $35 Boy’s Haircut $15+ Girl’s Haircut $20+ HAIRCARE GET IN TOUCH Keratin Treatments (905) 785-1100 Neckline Length $120+ Shoulder Length $150+ [email protected] Beauty & Hair Studio Shoulder Blade Length $175+ Waist Length $190+ Bellussalon.ca Hair Smoothing $210+ PERM SERVICE FOLLOW US SALON & BEAUTY SERVICES MENU Permanent Hairstyle Bellus Beauty & Hair Studio Short Hair Perm $100+ Medium Hair Perm $120+ @bellusmississauga Long Hair Perm $140+ Men’s Hair Perm $80+ @bellusstudio1 *HST not included. **Service charges may apply. HAIR COLOUR HIGHLIGHTS FACIALS WAXING LASHES & BROWS Regular / Ammonia-Free Half / Full Head / Ombré & Balayage Signature Facials & Treatments Waxing / Nufree Wax / Hard Wax Tinting & Lifting Advanced skincare treatments are also available. Root Touch-Up $35 / $45 Neckline Length $80+ / $90+ / — Eyebrows $10 / $12 / — Eyebrow Tinting $25 Shoulder Length $55+ / $65+ Shoulder Length $100+ / $110+ -

Waxing's a Hairy Issue for Tweens - LA Daily News Page 1 of 3
Waxing's a hairy issue for tweens - LA Daily News Page 1 of 3 Waxing's a hairy issue for I've turned people away." tweens Esser-Thorin and other aestheticians are increasingly being placed in the delicate role of educating mothers of tweens on when and where By Susan Abram, Staff Writer hair removal should and shouldn't be done. Article Last Updated: 08/25/2008 12:08:56 AM PDT In some cases, if a child's hair growth is abnormally advanced, aestheticians might agree to remove hair from between the brows or on the toes. But in some cases, they are being asked by moms to do more. And the trend is booming across the nation, partly thanks to a generation of mothers accustomed to primping at day spas. Monday's Online Poll Other parents simply want their child to fit in, Are parents who seek waxing for their preteen and if that means tweezing a child's unibrow or daughters going too far? eliminating heavy growth from their legs, a mother or a father could be willing. SHERMAN OAKS - In the world of intimate body waxing, Cynthia Esser-Thorin routinely rips the "Right around 2005, there was an influx and we hair follicles from the sensitive areas of the rich saw the numbers of teens visiting salons and and famous - and their wannabes - who come to spas increase by 25percent," said Bonnie Pink Cheeks, her Ventura Boulevard salon. Canavino, president of Amrit Organics, who also speaks on behalf of the American Association for But she draws the line when the client isn't old Aesticians. -

H Allen, Untitled ……………………………………………………… 18
HAIR Volume VI, Spring 2017 Hurford Center for the Arts and Humanities, Haverford College HAIR Volume VI, Spring 2017 Haverford College Margin is Haverford’s themed student-edited publication. Each issue features a topic marginalized in academic discourses, presenting sub- missions of critical essays, reviews, creative writing, visual media, and any other artifacts that critically or creatively engage the theme. We seek to publish the work of students, scholars, artists, musicians, and writers, both from within and outside of the Haverford community. [email protected] hav.to/margin haverford.edu/hcah Design by Duncan Cooper Cover: TKTKTKTKTK Sponsored by the John B. Hurford ’60 Center for the Arts and Humanities, Haverford College. TABLE OF CONTENTS Kaylynn Mayo, It’s My Hair (I Bought It) ……………………………………… 6 Jonathan Zelinger, Aristotle Didn’t Go to Medical School ………………… 8 Alina Wang, Rebirth ………………………………………………………… 12 Alina Wang, Medusa ………………………………………………………… 14 Késha Hollins, From the Roots ……………………………………………… 15 Chloe Wang, Personal Abyss …………………………………………………17 Alliyah Allen, Untitled ……………………………………………………… 18 Alliyah Allen, Untitled ……………………………………………………… 19 Isabella Siegel, Miss Wild …………………………………………………… 20 Nana Nieto, The Limit Does Not Exist ……………………………………… 22 Mercedes Davis, Recognizing the Beauty Within: Breaking Free From the Oppression of my Kinks and Curls ………………… 24 Alexandra Ben-Abba, Glass Haircut ……………………………………… 26 Lauren Sahlman, Barber Shop Moms ……………………………………… 28 Anonymous, Untitled ……………………………………………………… 29 Xiyuan Li, Untitled ………………………………………………………… 30 Glorín Colón, “Apollo’s Hair” ……………………………………………… 32 Essence Jackson-Jones, Untitled …………………………………………… 33 Essence Jackson-Jones, Untitled …………………………………………… 34 Essence Jackson-Jones, Untitled …………………………………………… 35 Contributors ……………………………………………………………… 36 LETTER FROM THE EDITOR Hair plays and has long played a critical role in people’s expression of self, polit- icized identities, and belonging. Traditionally, Han Chinese men and women alike wore their hair long, while in the U.S. -

Legs Bikini Arms Facial Features Body
VIVA MED AESTHETICS A FULL ARRAY OF SPA SERVICES TO “ T ENSION IS WHAT YOU THINK YOU SHOULD BE. RELAX REFRESH & REJUVENATE. R ELAXATION IS WHO YOU ARE.” Chinese proverb A Luxurious Experience Awaits Here at Viva Med we want you to look and feel your best! That’s why, LASER HAIR REMOVAL in addition to our concierge healthcare services, we also offer massage and aesthetic services Therapeutic Massage Relax, Rejuvenate, Refresh: Legs Skin Spot lightening Take a moment to feel and Lower $200 Body Sculpting Starters: look refreshed and younger Upper $250 Laser Hair Removal Facial Priced at consultation with our age defying Full $425 Chemical Peels Microderambrasion $80 treatments. Massage $60-$110 Aromatherapy Bikini Modest $175 French $200 Brazilian $250 Fractora: Nonsurgical Face Lift Arms Skin Resurfacing that combines microneedling with Under Arms $125 radiofrequency energy to bring back a youthful appearance. Arms $250 Great for antiaging, acne, acne scars, and other skin blemishes. Finger/Toes* $100 $500-$1000 Priced at Consultation Facial Features Body Contouring Lips $100 Body FX combines radio frequency energy and vacuum suction to Chin $100 reduce cellulite, tighten skin, and shrink fat deposits. Typically, it Neck $100 takes 4-6 sessions to achieve maximal results. Finally, a way to Combo: Lip/Chin $170 tighten the baby bulge, love handles, and thigh gaps! Combo: Chin/Neck $170 $250-$500 Priced at Consultation Unibrow $100 Beard $190 Votiva: Vaginal Rejuvenation Body Votiva with FormaV improves vaginal blood circulation and muscle Chest $300 relaxation and is an adjunct to Kegel exercises (tightening of the Full Back $325 muscles of the pelvic floor to increase muscle tone).