AP1B1 Rabbit Polyclonal Antibody – TA323221 | Origene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomic Aberrations Associated with Erlotinib Resistance in Non-Small Cell Lung Cancer Cells
ANTICANCER RESEARCH 33: 5223-5234 (2013) Genomic Aberrations Associated with Erlotinib Resistance in Non-small Cell Lung Cancer Cells MASAKUNI SERIZAWA1, TOSHIAKI TAKAHASHI2, NOBUYUKI YAMAMOTO2,3 and YASUHIRO KOH1 1Drug Discovery and Development Division, Shizuoka Cancer Center Research Institute, Sunto-gun, Shizuoka, Japan; 2Division of Thoracic Oncology, Shizuoka Cancer Center Hospital, Sunto-gun, Shizuoka, Japan; 3Third Department of Internal Medicine, Wakayama Medical University, Kimiidera, Wakayama, Japan Abstract. Background/Aim: Mechanisms of resistance to mutations develop resistance, usually within one year of epidermal growth factor receptor (EGFR)-tyrosine kinase treatment. Therefore, there is an urgent need to elucidate the inhibitors (TKIs) in non-small cell lung cancer (NSCLC) underlying mechanisms of resistance in such tumors to are not fully-understood. In this study we aimed to overcome this obstacle (11-14, 17, 24). Recent studies elucidate remaining unknown mechanisms using erlotinib- suggest that mechanisms of acquired resistance to EGFR- resistant NSCLC cells. Materials and Methods: We TKIs can be categorized into three groups: occurrence of performed array comparative genomic hybridization genetic alterations, activation of downstream pathways via (aCGH) to identify genomic aberrations associated with bypass signaling, and phenotypic transformation (15, 16, 21); EGFR-TKI resistance in erlotinib-resistant PC-9ER cells. therapeutic strategies to overcome these resistance Real-time polymerase chain reaction (PCR) and mechanisms are under development. However, although the immunoblot analyses were performed to confirm the results causes of acquired resistance to EGFR-TKIs have been of aCGH. Results: Among the five regions with copy investigated, in more than 30% of patients with acquired number gain detected in PC-9ER cells, we focused on resistance to EGFR-TKI treatment, the mechanisms remain 22q11.2-q12.1 including v-crk avian sarcoma virus CT10 unknown (15). -

University of California Santa Cruz Sample
UNIVERSITY OF CALIFORNIA SANTA CRUZ SAMPLE-SPECIFIC CANCER PATHWAY ANALYSIS USING PARADIGM A dissertation submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in BIOMOLECULAR ENGINEERING AND BIOINFORMATICS by Stephen C. Benz June 2012 The Dissertation of Stephen C. Benz is approved: Professor David Haussler, Chair Professor Joshua Stuart Professor Nader Pourmand Dean Tyrus Miller Vice Provost and Dean of Graduate Studies Copyright c by Stephen C. Benz 2012 Table of Contents List of Figures v List of Tables xi Abstract xii Dedication xiv Acknowledgments xv 1 Introduction 1 1.1 Identifying Genomic Alterations . 2 1.2 Pathway Analysis . 5 2 Methods to Integrate Cancer Genomics Data 10 2.1 UCSC Cancer Genomics Browser . 11 2.2 BioIntegrator . 16 3 Pathway Analysis Using PARADIGM 20 3.1 Method . 21 3.2 Comparisons . 26 3.2.1 Distinguishing True Networks From Decoys . 27 3.2.2 Tumor versus Normal - Pathways associated with Ovarian Cancer 29 3.2.3 Differentially Regulated Pathways in ER+ve vs ER-ve breast can- cers . 36 3.2.4 Therapy response prediction using pathways (Platinum Free In- terval in Ovarian Cancer) . 38 3.3 Unsupervised Stratification of Cancer Patients by Pathway Activities . 42 4 SuperPathway - A Global Pathway Model for Cancer 51 4.1 SuperPathway in Ovarian Cancer . 55 4.2 SuperPathway in Breast Cancer . 61 iii 4.2.1 Chin-Naderi Cohort . 61 4.2.2 TCGA Breast Cancer . 63 4.3 Cross-Cancer SuperPathway . 67 5 Pathway Analysis of Drug Effects 74 5.1 SuperPathway on Breast Cell Lines . -

A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection
CLINICAL RESEARCH www.jasn.org A Peripheral Blood Gene Expression Signature to Diagnose Subclinical Acute Rejection Weijia Zhang,1 Zhengzi Yi,1 Karen L. Keung,2 Huimin Shang,3 Chengguo Wei,1 Paolo Cravedi,1 Zeguo Sun,1 Caixia Xi,1 Christopher Woytovich,1 Samira Farouk,1 Weiqing Huang,1 Khadija Banu,1 Lorenzo Gallon,4 Ciara N. Magee,5 Nader Najafian,5 Milagros Samaniego,6 Arjang Djamali ,7 Stephen I. Alexander,2 Ivy A. Rosales,8 Rex Neal Smith,8 Jenny Xiang,3 Evelyne Lerut,9 Dirk Kuypers,10,11 Maarten Naesens ,10,11 Philip J. O’Connell,2 Robert Colvin,8 Madhav C. Menon,1 and Barbara Murphy1 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background In kidney transplant recipients, surveillance biopsies can reveal, despite stable graft function, histologic features of acute rejection and borderline changes that are associated with undesirable graft outcomes. Noninvasive biomarkers of subclinical acute rejection are needed to avoid the risks and costs associated with repeated biopsies. Methods We examined subclinical histologic and functional changes in kidney transplant recipients from the prospective Genomics of Chronic Allograft Rejection (GoCAR) study who underwent surveillance biopsies over 2 years, identifying those with subclinical or borderline acute cellular rejection (ACR) at 3 months (ACR-3) post-transplant. We performed RNA sequencing on whole blood collected from 88 indi- viduals at the time of 3-month surveillance biopsy to identify transcripts associated with ACR-3, developed a novel sequencing-based targeted expression assay, and validated this gene signature in an independent cohort. -

Clathrin Heavy and Light Chain Isoforms Originated by Independent Mechanisms of Gene Duplication During Chordate Evolution
Clathrin heavy and light chain isoforms originated by independent mechanisms of gene duplication during chordate evolution Diane E. Wakeham†‡, Laurent Abi-Rached‡§, Mhairi C. Towler†¶, Jeremy D. Wilbur†ʈ, Peter Parham§, and Frances M. Brodsky†,†† †The G. W. Hooper Foundation and Departments of Biopharmaceutical Sciences, Pharmaceutical Chemistry, and Microbiology and Immunology and ʈBiophysics Program, University of California, San Francisco, CA 94143-0552; and §Department of Structural Biology and Department of Microbiology and Immunology, Stanford University, Stanford, CA 94305 Communicated by J. Michael Bishop, University of California, San Francisco, CA, March 22, 2005 (received for review January 14, 2005) In humans, there are two isoforms each of clathrin heavy chain biogenesis. CHC17 is expressed ubiquitously in vertebrate tissues (CHC17 and CHC22) and light chain (LCa and LCb) subunits, all and a functional orthologue is present in all eukaryotic organ- encoded by separate genes. CHC17 forms the ubiquitous clathrin- isms analyzed. The CHC22 isoform is highly expressed in human coated vesicles that mediate membrane traffic. CHC22 is implicated skeletal muscle, with a low level detected in other tissues. CHC22 in specialized membrane organization in skeletal muscle. CHC17 is is concentrated at neuromuscular and myotendinous junctions, bound and regulated by LCa and LCb, whereas CHC22 does not and its expression increases during myogenesis and muscle functionally interact with either light chain. The imbalanced inter- regeneration (10, 11). Thus, CHC22 appears to have a role actions between clathrin subunit isoforms suggest a distinct evo- distinct from CHC17 in specialized muscle membrane organi- lutionary history for each isoform pair. Phylogenetic and sequence zation. CHC17 and CHC22 have a remarkably high protein sequence identity (85%), despite their evident differences in analysis placed both heavy and light chain gene duplications function. -

Supplementary Information – Postema Et Al., the Genetics of Situs Inversus Totalis Without Primary Ciliary Dyskinesia
1 Supplementary information – Postema et al., The genetics of situs inversus totalis without primary ciliary dyskinesia Table of Contents: Supplementary Methods 2 Supplementary Results 5 Supplementary References 6 Supplementary Tables and Figures Table S1. Subject characteristics 9 Table S2. Inbreeding coefficients per subject 10 Figure S1. Multidimensional scaling to capture overall genomic diversity 11 among the 30 study samples Table S3. Significantly enriched gene-sets under a recessive mutation model 12 Table S4. Broader list of candidate genes, and the sources that led to their 13 inclusion Table S5. Potential recessive and X-linked mutations in the unsolved cases 15 Table S6. Potential mutations in the unsolved cases, dominant model 22 2 1.0 Supplementary Methods 1.1 Participants Fifteen people with radiologically documented SIT, including nine without PCD and six with Kartagener syndrome, and 15 healthy controls matched for age, sex, education and handedness, were recruited from Ghent University Hospital and Middelheim Hospital Antwerp. Details about the recruitment and selection procedure have been described elsewhere (1). Briefly, among the 15 people with radiologically documented SIT, those who had symptoms reminiscent of PCD, or who were formally diagnosed with PCD according to their medical record, were categorized as having Kartagener syndrome. Those who had no reported symptoms or formal diagnosis of PCD were assigned to the non-PCD SIT group. Handedness was assessed using the Edinburgh Handedness Inventory (EHI) (2). Tables 1 and S1 give overviews of the participants and their characteristics. Note that one non-PCD SIT subject reported being forced to switch from left- to right-handedness in childhood, in which case five out of nine of the non-PCD SIT cases are naturally left-handed (Table 1, Table S1). -

Comparative Proteomic Analysis Identifies Epha2 As a Specific Cell
International Journal of Molecular Sciences Article Comparative Proteomic Analysis Identifies EphA2 as a Specific Cell Surface Marker for Wharton’s Jelly-Derived Mesenchymal Stem Cells Ashraf Al Madhoun 1,2,* , Sulaiman K. Marafie 3 , Dania Haddad 2, Motasem Melhem 2, Mohamed Abu-Farha 3 , Hamad Ali 2,4 , Sardar Sindhu 1 , Maher Atari 5 and Fahd Al-Mulla 2 1 Department of Animal and Imaging Core Facilities, Dasman Diabetes Institute, Dasman 15462, Kuwait; [email protected] 2 Department of Genetics and Bioinformatics, Dasman Diabetes Institute, Dasman 15462, Kuwait; [email protected] (D.H.); [email protected] (M.M.); [email protected] (H.A.); [email protected] (F.A.-M.) 3 Department of Biochemistry and Molecular Biology, Dasman Diabetes Institute, Dasman 15462, Kuwait; sulaiman.marafi[email protected] (S.K.M.); [email protected] (M.A.-F.) 4 Department of Medical Laboratory Sciences, Faculty of Allied Health Sciences, Health Sciences Center (HSC), Kuwait University, Jabriya 046302, Kuwait 5 Medical-Surgical Pathology Department, Regenerative Medicine Research Institute, Universitat Internacional de Catalunya, 08195 Barcelona, Spain; [email protected] * Correspondence: [email protected] Received: 28 June 2020; Accepted: 1 September 2020; Published: 3 September 2020 Abstract: Wharton’s jelly-derived mesenchymal stem cells (WJ-MSCs) are a valuable tool in stem cell research due to their high proliferation rate, multi-lineage differentiation potential, and immunotolerance properties. However, fibroblast impurity during WJ-MSCs isolation is unavoidable because of morphological similarities and shared surface markers. Here, a proteomic approach was employed to identify specific proteins differentially expressed by WJ-MSCs in comparison to those by neonatal foreskin and adult skin fibroblasts (NFFs and ASFs, respectively). -

Transforming Growth Factor ß1-Mediated Functional Inhibition Of
Myelodysplastic Syndromes SUPPLEMENTARY APPENDIX Transforming growth factor 1- mediated functional inhibition of mesenchymal stromal celβls in myelodysplastic syndromes and acute myeloid leukemia Stefanie Geyh, 1* Manuel Rodríguez-Paredes, 1,2 * Paul Jäger, 1 Annemarie Koch, 1 Felix Bormann, 2 Julian Gutekunst, 2 Christoph Zilkens, 3 Ulrich Germing, 1 Guido Kobbe, 1 Frank Lyko, 2 Rainer Haas 1 and Thomas Schroeder 1 1Department of Hematology, Oncology and Clinical Immunology, University of Duesseldorf, Medical Faculty; 2Division of Epigenetics, DKFZ- ZMBH Alliance, German Cancer Research Center, Heidelberg and 3Department of Orthopedic Surgery, University of Duesseldorf, Medical Faculty, Germany *SG and MR-P contributed equally to this work. ©2018 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.186734 Received: December 19, 2017. Accepted: May 14, 2018. Pre-published: May 17, 2018. Correspondence: [email protected] Figure S1 Downregulated genes Downregulated genes Upregulated Figure S1. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 9 RCMD-, RAEB- and AML-derived MSC samples. Color scale depicts the rlog-transformed FPKM values for each gene and every sample. Figure S2 Downregulated genes Downregulated genes Upregulated Figure S2. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 3 RCMD, RAEB and AML MSC samples, respectively. Color scales depict the rlog-transformed FPKM values for each gene and every sample. Figure S3 A. B. 0.0015 *** ** <-3 -2 0.0010 RCMD RAEB AML -1 0 1 0.0005 Log2FC LTF 2 CCL26/GAPDH INHBB >3 0.0000 TGFB2 y S h D ML M A ealt ll LTF H a EGF 0.003 *** ** INHBB TGFB2 0.002 INHBB IGFBP7 0.001 GDF11 LIF/GAPDH BMP1 0.000 y L th M TNFSF12 l A FGF13 ea ll MDS H a FGF13 0.0015 * TNFSF10 TNFSF10 0.0010 0.0005 SPP1/GAPDH 0.0000 y th l AML ea H all MDS Figure S3. -

Cilia and Ciliopathies in Congenital Heart Disease
Downloaded from http://cshperspectives.cshlp.org/ on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Cilia and Ciliopathies in Congenital Heart Disease Nikolai T. Klena, Brian C. Gibbs, and Cecilia W. Lo Department of Developmental Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15201 Correspondence: [email protected] A central role for cilia in congenital heart disease (CHD) was recently identified in a large- scale mouse mutagenesis screen. Although the screen was phenotype-driven, the majority of genes recovered were cilia-related, suggesting that cilia play a central role in CHD patho- genesis. This partly reflects the role of cilia as a hub for cell signaling pathways regulating cardiovascular development. Consistent with this, manycilia-transduced cell signaling genes were also recovered, and genes regulating vesicular trafficking, a pathway essential for cilio- genesis and cell signaling. Interestingly, among CHD-cilia genes recovered, some regulate left–right patterning, indicating cardiac left–right asymmetry disturbance may play signifi- cant roles in CHD pathogenesis. Clinically, CHD patients show a high prevalence of ciliary dysfunction and show enrichment for de novo mutations in cilia-related pathways. Combined with the mouse findings, this would suggest CHD may be a new class of ciliopathy. ongenital heart disease (CHD) is one of the shown to have a high recurrence risk, with fa- Cmost common birth defects, found in an milial clustering indicating a genetic contribu- estimated 1% of live births (Hoffman and tion (Gill et al. 2003; Oyen et al. 2009). The Kaplan 2002). With advances in surgical pallia- identification of the genetic causes of CHD tion, most patients with CHD now survive their may provide mechanistic insights that can critical heart disease such that currently there help stratify patients for guiding the therapeutic are more adults with CHD than infants born management of their clinical care. -

Product Size GOT1 P00504 F CAAGCTGT
Table S1. List of primer sequences for RT-qPCR. Gene Product Uniprot ID F/R Sequence(5’-3’) name size GOT1 P00504 F CAAGCTGTCAAGCTGCTGTC 71 R CGTGGAGGAAAGCTAGCAAC OGDHL E1BTL0 F CCCTTCTCACTTGGAAGCAG 81 R CCTGCAGTATCCCCTCGATA UGT2A1 F1NMB3 F GGAGCAAAGCACTTGAGACC 93 R GGCTGCACAGATGAACAAGA GART P21872 F GGAGATGGCTCGGACATTTA 90 R TTCTGCACATCCTTGAGCAC GSTT1L E1BUB6 F GTGCTACCGAGGAGCTGAAC 105 R CTACGAGGTCTGCCAAGGAG IARS Q5ZKA2 F GACAGGTTTCCTGGCATTGT 148 R GGGCTTGATGAACAACACCT RARS Q5ZM11 F TCATTGCTCACCTGCAAGAC 146 R CAGCACCACACATTGGTAGG GSS F1NLE4 F ACTGGATGTGGGTGAAGAGG 89 R CTCCTTCTCGCTGTGGTTTC CYP2D6 F1NJG4 F AGGAGAAAGGAGGCAGAAGC 113 R TGTTGCTCCAAGATGACAGC GAPDH P00356 F GACGTGCAGCAGGAACACTA 112 R CTTGGACTTTGCCAGAGAGG Table S2. List of differentially expressed proteins during chronic heat stress. score name Description MW PI CC CH Down regulated by chronic heat stress A2M Uncharacterized protein 158 1 0.35 6.62 A2ML4 Uncharacterized protein 163 1 0.09 6.37 ABCA8 Uncharacterized protein 185 1 0.43 7.08 ABCB1 Uncharacterized protein 152 1 0.47 8.43 ACOX2 Cluster of Acyl-coenzyme A oxidase 75 1 0.21 8 ACTN1 Alpha-actinin-1 102 1 0.37 5.55 ALDOC Cluster of Fructose-bisphosphate aldolase 39 1 0.5 6.64 AMDHD1 Cluster of Uncharacterized protein 37 1 0.04 6.76 AMT Aminomethyltransferase, mitochondrial 42 1 0.29 9.14 AP1B1 AP complex subunit beta 103 1 0.15 5.16 APOA1BP NAD(P)H-hydrate epimerase 32 1 0.4 8.62 ARPC1A Actin-related protein 2/3 complex subunit 42 1 0.34 8.31 ASS1 Argininosuccinate synthase 47 1 0.04 6.67 ATP2A2 Cluster of Calcium-transporting -

Aberrant Alternative Splicing and Extracellular Matrix Gene Expression in Mouse
Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Hongqing Du1, Melissa S. Cline1, Robert J. Osborne2, Daniel L. Tuttle3, Tyson A. Clark4, John Paul Donohue1, Megan P. Hall1, Lily Shiue1, Maurice S. Swanson3, Charles A. Thornton2, and Manuel Ares, Jr.1* 1RNA Center, Department of Molecular, Cell and Developmental Biology, Sinsheimer Labs, University of California, Santa Cruz, California 95064 USA 2Neuromuscular Disease Center, Department of Neurology, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642 USA 3Department of Molecular Genetics & Microbiology, University of Florida, College of Medicine, Gainesville, FL 32610 USA 4Affymetrix Inc, Santa Clara, CA 95051 USA SUPPLEMENTARY FIGURES AND TABLES Nature Structural & Molecular Biology: doi:10.1038/nsmb.1720 Nature Structural & Molecular Biology: doi:10.1038/nsmb.1720 Supplementary Figure 1 Validation and comparison of mis-spliced events in quadriceps samples of HSALR and MBNL1ΔE3/ΔE3 mice. RT-PCR fragments were separated on 2.5% agarose gel. The mis- splicing events validated by RT-PCR were classified (a-c): 28 RT-PCR validations of mis-splicing cassette exon events predicted by splicing microarray to be altered in both HSALR and MBNL1ΔE3/ΔE3 mice (a); 4 mis-spliced cassette exon events altered only in MBNL ΔE3/ΔE3 mice (b); 1 mis-splicing cassette exon event altered only in HSALR mice (c). (d) Comparison of separation score of altered splicing events predicted by RT-PCR in both HSALR mice and MBNL1ΔE3/ΔE3 mice (R2 = 0.88). Separation score for RT-PCR data were calculated using amounts determined by the Bioanalyzer. Nature Structural & Molecular Biology: doi:10.1038/nsmb.1720 Supplementary Figure 2 Mapping binding motifs for other splicing factors. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -
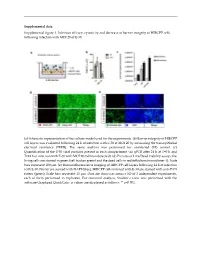
Supplemental Figure 1. Infection Efficacy, Cytoxicity and Decrease in Barrier Integrity of HIBCPP Cells Following Infection with MOI 20 of E-30
Supplemental data: Supplemental figure 1. Infection efficacy, cytoxicity and decrease in barrier integrity of HIBCPP cells following infection with MOI 20 of E-30. (a) Schematic representation of the culture model used for the experiments. (b) Barrier integrity of HIBCPP cell layers was evaluated following 24 h of infection with E-30 at MOI 20 by measuring the transepithelial electrical resistance (TEER). The same analysis was performed for uninfected (UI) control. (c) Quantification of the E-30 viral particles present in each compartment via qPCR after 24 h at T=0 h and T=24 h of infection with E-30 with MOI 20 (nd=non detected). (d) Pictures of Live/Dead viability assays; the living cells are stained in green (cell tracker green) and the dead cells in red (ethidium homodimer-1). Scale bars represent 100 µm. (e) Immunofluorescence imaging of HIBCPP cell layers following 24 h of infection with E-30. Nuclei are stained with DAPI (blue), HIBCPP cells infected with E-30 are stained with anti-PAN entero (green). Scale bars represent 15 µm. Data are shown as mean ± SD of 3 independent experiments, each of them performed in triplicates. For statistical analysis, Student’s t-test was performed with the software Graphpad QuickCalcs. p values are displayed as follows: ** p<0.001. Supplemental table 1. Up-regulated proteins in the lipid raft of HIBCPP cells following 24 h of E-30 infection at MOI 20. Proteins were identified as significantly changed in abundancy, when they exceeded a t-test difference > 1.0. Results shown are the comparison of 3 different independent proteomic experiments.