Cicer Arietinum L.
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Zn-Use Efficiency for Optimization of Symbiotic Nitrogen Fixation in Chickpea (Cicer Arietinum L.)
Turkish Journal of Botany Turk J Bot (2017) 41: 423-441 http://journals.tubitak.gov.tr/botany/ © TÜBİTAK Review Article doi:10.3906/bot-1610-6 Zn-use efficiency for optimization of symbiotic nitrogen fixation in chickpea (Cicer arietinum L.) Igor S. KRYVORUCHKO* Department of Bioengineering, Faculty of Engineering and Architecture, Kafkas University, Kars, Turkey Received: 04.10.2016 Accepted/Published Online: 28.05.2017 Final Version: 28.09.2017 Abstract: Zn deficiency is widespread in traditional areas of chickpea cultivation worldwide. It limits chickpea productivity and causes significant losses to the economies of the world’s largest chickpea exporters. This review may be of interest to researchers who would like to contribute to the improvement of chickpea cultivation on Zn-depleted soils in an environmentally sustainable manner, namely via identification of genotypes with superior symbiotic performance under Zn-limited conditions. The primary aim of the current work is to familiarize the readers with the biology and symbiotic characteristics of chickpea, and also to provide the necessary background on Zn as an essential nutrient for symbiotic nitrogen fixation (SNF). Special attention has been paid to the choice of rhizobial strains compatible with chickpea. Strains that can serve as an inoculum for simultaneous analysis of many genetically diverse chickpea lines have been suggested. The genotypes listed in this work can be good starting material for identification of chickpea lineages useful for unraveling the molecular basis of Zn-use efficiency, SNF efficiency, or both. Key words: Chickpea, Cicer arietinum, symbiotic nitrogen fixation, nodulation, rhizobia, zinc, micronutrient deficiency 1. Introduction on its position in international trade. -

Vernalization Response in Chickpea Is Controlled by a Major QTL
Euphytica (2016) 207:453–461 DOI 10.1007/s10681-015-1571-4 Vernalization response in chickpea is controlled by a major QTL Srinivasan Samineni . Suresh Kamatam . Mahendar Thudi . Rajeev K. Varshney . Pooran M. Gaur Received: 11 June 2015 / Accepted: 27 September 2015 / Published online: 7 October 2015 Ó Springer Science+Business Media Dordrecht 2015 Abstract Wild Cicer species are known to be more response genes. Among 84 flowering related genes responsive to vernalization (induce early flowering present in this region, Ca_06280 related to MADS box when exposed to low temperatures) than the cultivated genes, was reported to play important role in vernal- chickpea. This study was aimed at molecular mapping ization in cereals. Understanding the genetic control of of vernalization response quantitative trait loci (QTLs) vernalization response in chickpea will help in in chickpea. An interspecific recombinant inbred line exploitation of wild Cicer species in chickpea (RIL) population [ICC 4958 (Cicer arietinum) 9 PI improvement. 489777 (Cicer reticulatum)] was phenotyped for vernalization response for two consecutive years Keywords Flowering time Á Cold treatment Á QTL Á (2009–2010 and 2010–2011) under field conditions. Linkage map A linkage map already available for this population was used for QTL analysis. A major QTL contributing 55 % of phenotypic variation for vernalization Introduction response trait was identified on LG 3 at LOD score of 27. The simple sequence repeat (SSR) markers Chickpea (Cicer arietinum L.) is an important cool- TA64 and CaM1515 were flanking the QTL which season food legume crop mainly grown in the arid and spans a distance of 22 cM. -

Cicer Arietinum L.)
agronomy Article Integration of Extra-Large-Seeded and Double-Podded Traits in Chickpea (Cicer arietinum L.) Kamile Gul Kivrak 1, Tuba Eker 1, Hatice Sari 1 , Duygu Sari 1, Kadir Akan 2, Bilal Aydinoglu 1, Mursel Catal 3 and Cengiz Toker 1,* 1 Department of Field Crops, Faculty of Agriculture, Akdeniz University, 07070 Antalya, Turkey; [email protected] (K.G.K.); [email protected] (T.E.); [email protected] (H.S.); [email protected] (D.S.); [email protected] (B.A.) 2 Department of Plant Protection, Faculty of Agriculture, Kirsehir Ahi Evran University, 40200 Kirsehir, Turkey; [email protected] 3 Department of Plant Protection, Faculty of Agriculture, Akdeniz University, 07070 Antalya, Turkey; [email protected] * Correspondence: [email protected]; Tel.: +90-242-310-24-21 or +90-537-543-10-37 Received: 1 June 2020; Accepted: 22 June 2020; Published: 24 June 2020 Abstract: A large seed size in the kabuli chickpea (Cicer arietinum L.) is important in the market not only due to its high price but also for its superior seedling vigor. The double-podded chickpea has a considerable yield and stability advantage over the single-podded chickpea. The study aimed at (i) integrating extra-large-seeded and double-podded traits in the kabuli chickpea, (ii) increasing variation by transgressive segregations and (iii) estimating the heritability of the 100-seed weight along with important agro-morphological traits in F2 and F3 populations. For these objectives, the large-seeded chickpea, Sierra, having a single pod and unifoliolate leaves, was crossed with the small-seeded CA 2969, having double pods and imparipinnate leaves. -

Investigation of the Development of Root Lesion Nematodes, Pratylenchus Spp
Türk. entomol. derg., 2021, 45 (1): 23-31 ISSN 1010-6960 DOI: http://dx.doi.org/10.16970/entoted.753614 E-ISSN 2536-491X Original article (Orijinal araştırma) Investigation of the development of root lesion nematodes, Pratylenchus spp. (Tylenchida: Pratylenchidae) in three chickpea cultivars Kök lezyon nematodlarının, Pratylenchus spp. (Tylenchida: Pratylenchidae) üç nohut çeşidinde gelişmesinin incelenmesi İrem AYAZ1 Ece B. KASAPOĞLU ULUDAMAR1* Tohid BEHMAND1 İbrahim Halil ELEKCİOĞLU1 Abstract In this study, penetration, population changes and reproduction rates of root lesion nematodes, Pratylenchus neglectus (Rensch, 1924), Pratylenchus penetrans (Cobb, 1917) and Pratylenchus thornei Sher & Allen, 1953 (Tylenchida: Pratylenchidae), at 3, 7, 14, 21, 28, 35, 42, 49 and 56 d after inoculation in chickpea Bari 2, Bari 3 (Cicer reticulatum Ladiz) and Cermi [Cicer echinospermum P.H.Davis (Fabales: Fabaceae)] were assessed in a controlled environment room in 2018-2019. No juveniles were observed in the roots in the first 3 d after inoculation. Although, population density of P. thornei reached the highest in Cermi (21 d), Bari 3 (42 d) and the lowest observed on Bari 2. Pratylenchus neglectus reached the highest population density in Bari 3 and Cermi on day 28. The population density of P. neglectus was the lowest in Bari 2. Also, population density of P. penetrans reached the highest in Bari 3 cultivar within 49 d, similar to P. thornei, whereas Bari 2 and Cermi had low population densities during the entire experimental period. Keywords: -

Chapter 4. Chickpea
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by ICRISAT Open Access Repository 4 Chickpea Shivali Sharma, Hari D. Upadhyaya, Manish Roorkiwal, Rajeev K. Varshney and C.L. Laxmipathi Gowda International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Hyderabad, India 4.1 Introduction Chickpea (Cicer arietinum L.) is a self-pollinated true diploid (2n=2x=16) cool season leguminous crop that ranks second among food grain legumes in the world after common bean (FAOSTAT, 2011). It is grown in a wide range of environments in over 50 countries in subtropical and temperate regions of the world, mainly in the Indian subcontinent, West Asia, North Africa, the Americas and Australia (FAOSTAT, 2011). Based on seed shape, size and colour, two distinct forms of cul- tivated chickpea are known (Cubero, 1975); namely, the desi type, characterized mostly by pink flowers, angular. brown, small seeds with a high percentage of fibre, primarily grown in South Asia and Africa; and kabuli type, having white flowers and owl-head-shaped, beige, large seeds with a low percentage of fibre, grown in Mediterranean countries. A third type, designated as intermediate or pea-shaped, is characterized by medium to small size and round, pea-shaped seeds. Kabuli types are grown in about two-thirds of chickpea-growing countries, but desi type predominates in chickpea production and accounts for about 85%, while kabuli accounts for about 15% of the world chickpea production. It is grown primarily for its protein-rich seeds. In addition, chickpea seeds are also rich in minerals (calcium, potassium, phosphorus, magnesium, iron and zinc), fibre, unsaturated fatty acids, and β-carotene (Jukanti, Gaur, Gowda, & Chibbar, 2012). -
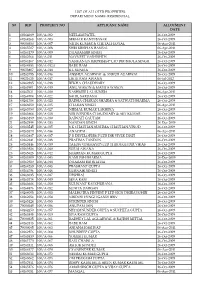
S# Rid Property No Applicant Name Allotment Date 1
LIST OF ALLOTED PROPERTIES DEPARTMENT NAME- RESIDENTIAL S# RID PROPERTY NO APPLICANT NAME ALLOTMENT DATE 1 60264689 100/A-002 NEELAM PATEL 26-Oct-2009 2 60264268 100/A-003 SEKHAR KANTI BASAK 26-Oct-2009 3 90000350 100/A-007 NITIN KUMAR & CHETALI GOYAL 06-Apr-2011 4 60265507 100/A-008 SHRI KRISHAN BANSAL 06-Apr-2011 5 60264179 100/A-009 DHARAMBIR SINGH 26-Oct-2009 6 60262564 100/A-011 NAVNEET VASHISHTH 26-Oct-2009 7 60264167 100/A-012 YASIKAN ENTERPRISES P LTD DIR BHOLA SINGH 26-Oct-2009 8 60264553 100/A-012A BABU RAM 26-Oct-2009 9 90023807 100/A-014 K L SUNEJA 26-Oct-2009 10 60265795 100/A-016 ANSHUL AGARWAL & ANKUR AGARWAL 26-Oct-2009 11 90021623 100/A-017 LIKHI RAM AWANA 06-Jul-2012 12 60262985 100/A-018 REKHA CHAUDHARY 26-Oct-2009 13 60263901 100/A-019 ANIL WASON & MAMTA WASON 26-Oct-2009 14 60267621 100/A-020 KASHMIRI LAL SUNEJA 06-Apr-2011 15 60264704 100/A-022 SAHIL SARDANA 26-Oct-2009 16 60261786 100/A-023 RADHA CHARAN SHARMA & SATWATI SHARMA 26-Oct-2009 17 60266050 100/A-025 CHARAN SINGH 06-Apr-2011 18 60263750 100/A-027 NIRMAL KUMAR LAKHINA 26-Oct-2009 19 60263868 100/A-028 VISHVENDRA CHAUDHARY & AJIT KUMAR 26-Oct-2009 20 60263189 100/A-030 RAJWATI GAUTAM 26-Oct-2009 21 60262994 100/A-033 NANDAN SINGH 26-Dec-2009 22 60263545 100/A-035 S K CHAUHAN,SUSHMA CHAUHAN,VINOD 26-Oct-2009 23 60265570 100/A-036 ANAGPAL 06-Apr-2011 24 60262887 100/A-037 P R DEVELOPERS P LTD DIR VIVEK DIXIT 26-Oct-2009 25 60262241 100/A-038 PRATIMA TANDON 26-Oct-2009 26 60263846 100/A-039 TALON VINIMAY(P) LTD THROUGH DIR.VIKAS 26-Oct-2009 27 60263926 100/A-040 -

Chickpea 3 Sarvjeet Singh , Inderjit Singh , Karan Kapoor , P
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by ICRISAT Open Access Repository Chickpea 3 Sarvjeet Singh , Inderjit Singh , Karan Kapoor , P. M. Gaur , S. K. Chaturvedi , N. P. Singh , and J. S. Sandhu Abstract The narrow genetic base of cultivated chickpea warrants systematic collection, documentation and evaluation of chickpea germplasm and particularly wild Cicer species for effective and effi cient use in chickpea breeding programmes. Limiting factors to crop production, possible solutions and ways to overcome them, importance of wild relatives and barriers to alien gene introgression and strategies to overcome them and traits for base broadening have been discussed. It has been clearly demonstrated that resistance to major biotic and abiotic stresses can be successfully introgressed from the primary gene pool comprising progenitor species. However, many desirable traits including high degree of resistance to multiple stresses that are present in the species belonging to secondary and tertiary gene pools can also be introgressed by using special techniques to overcome pre- and post-fertilization barriers. Besides resistance to various biotic and abiotic stresses, the yield QTLs have also been introgressed from wild Cicer species to cultivated varieties. Status and importance of molecular markers, genome mapping and genomic tools for chickpea improvement are elaborated. Because of major genes for various biotic and abiotic stresses, the transfer of agronomically important traits into elite cultivars has been made easy and practical through marker-assisted selection and marker-assisted backcross. The usefulness of molecular markers such as SSR and SNP for the construction of high-density genetic maps of chickpea and for the identifi cation of genes/QTLs for stress resistance, quality and yield contributing traits has also been discussed. -

The Use of Cicer Reticulatum L. for Genetic Improvement of Cultivated Chickpea
The use of Cicer reticulatum L. for genetic improvement of cultivated chickpea A Thesis Submitted to the College of Graduate and Postdoctoral Studies In Partial Fulfillment of the Requirements For the Degree of Master of Science In the Department of Plant Sciences University of Saskatchewan Saskatoon By Md. Waliur Rahman © Copyright Md. Waliur Rahman, April 2020. All rights reserved. PERMISSION TO USE In presenting this thesis in partial fulfillment of the requirements for a Postgraduate degree from the University of Saskatchewan, I agree that the Libraries of this University may make it freely available for inspection. I further agree that permission for copying this thesis in any manner, in whole or in part, for scholarly purposes may be granted by the professor or professors who supervised my thesis work, or in their absence, by the Head of the Department of Plant Sciences or the Dean of the College of Agriculture and Bioresources. It is understood that any copying or publication or use of the thesis, in whole or in part, for financial gain shall not be allowed without my written permission. It is also understood that due recognition shall be given to me and to the University of Saskatchewan in any scholarly use which may be made of any material in my thesis. Requests for permission to copy or to make other uses of materials in this thesis, in whole or in part, should be addressed to: Head Department of Plant Sciences University of Saskatchewan Saskatoon, Saskatchewan S7N 5A8 Canada Or Dean College of Graduate and Postdoctoral Studies University of Saskatchewan 116 Thorvaldson Building, 110 Science Place Saskatoon, Saskatchewan S7N 5C9 Canada i DISCLAIMER Reference in this thesis to any specific commercial products, process, or service by trade name, trademark, manufacturer, or otherwise, does not constitute or imply its endorsement, recommendation, or favouring by the University of Saskatchewan. -

Chickpea 1 Chickpea
Chickpea 1 Chickpea Chickpea Varieties Left, Bengal (Indian); right, European Scientific classification Kingdom: Plantae (unranked): Angiosperms (unranked): Eudicots (unranked): Rosids Order: Fabales Family: Fabaceae Genus: Cicer Species: C. arietinum Binomial name Cicer arietinum L. The chickpea (Cicer arietinum) is a legume of the family Fabaceae, subfamily Faboideae. Its seeds are high in protein. It is one of the earliest cultivated legumes: 7,500-year-old remains have been found in the Middle East.[1] Other common names for the species include garbanzo bean, ceci bean, chana, sanagalu, Gonzo Bean and Bengal gram. Etymology The name "chickpea" traces back through the French chiche to cicer, Latin for ‘chickpea’ (from which the Roman cognomen Cicero was taken). The Oxford English Dictionary lists a 1548 citation that reads, "Cicer may be named in English Cich, or ciche pease, after the Frenche tonge." The dictionary cites "Chick-pea" in the mid-18th century; the original word in English taken directly from French was chich, found in print in English in 1388 and became obsolete in the 18th century. The word garbanzo came to English as "calavance" in the 17th century, from Old Spanish (perhaps influenced by Old Spanish garroba or algarroba), though it came to refer to a variety of other beans (cf. Calavance). The Portuguese (?) arvanço has suggested to some that the origin of the word garbanzo is in the Greek erebinthos.[2] But the Oxford English Dictionary notes that some scholars doubt this; it also mentions a possible origination in the word Chickpea 2 garbantzu, from Basque — a non-Indo-European tongue — in which it is a compound of garau, seed + antzu, dry. -

Behsdabstractbook.Pdf
CONTENTS Messages Committees BEHSD – 2018 Organizers Detailed Scientific Program Abstracts of Plenary and Invited Lectures Abstracts of Short Oral Presentations Poster Abstracts About BRSI Sponsor Message from Professor Alok Dhawan, Director, CSIR-IITR Message from Dr D Parmar, Chairman, BEHSD-2018 Message from Professor TP Singh, President, BRSI Message from Professor Ashok Pandey, General Chair, BEHSD-2018 Bio-Innovation for Environmental and Health Sustainable Developments (BEHSD-2018) Bio-Innovation for Environmental and Health Sustainable Developments (BEHSD-2018) November 27-28, 2018 Organizing Committee Patron: Professor Alok Dhawan General Chairman: Professor Ashok Pandey Conference Chairman- Dr D Parmar Convener: Dr N Manickam Co-Conveners: Dr Kausar Ansari, Dr R Parthasarathi Treasurer- Dr K C Khulbe COA, CoFA, Sr. FAO, SPO, AO Committees and Members Fund Raising Catering and Refreshment Dr Poonam Kakkar (Chair Person) Dr K C Khulbe (Chairman) Dr Ratan Singh Ray (Convener) Dr Sanjay Yadav (Convener) Dr Sanjay Yadav Dr Sandeep Sharma Dr Sheelendra Pratap Singh Dr Ram Narayan Mr Laxmi Kant (Co-Convener) Scientific Session & Website, Publicity and Press Publication Audio visual & photography Dr K Chowdhuri (Chairman) Dr Yogeshwer Shukla (Chairman) Dr Ravi Ram Kristipati (Convener) Mr Nikhil Garg (Convener) Dr Akshay Dwarakanath Dr Akshay Dwarakanath Dr Amit Kumar Mr C M Tiwari Dr Vikas Srivatsava Mr Ali Kausar Dr R Rajagopal Mr Nasir Naqvi Dr Neeraj Satija Mr Shyam Kumar Pal Mr Sarfraj Ahmed Venue/stage Arrangement Transport Dr Alok Pandey (Chairman) Dr Ratan Singh Ray (Chairman) Dr Kausar M Ansari (Convener) Er A H Khan (Convener) Mr Yogendra Singh Dr Dhirendra Singh Mr R K Upadhyay Dr Anjeneya Dr Somendu Roy Mr Sandeep Negi Mr. -

M. Imran Kozgar Mutation Breeding in Chickpea: Perspectives and Prospects for Food Security
M. Imran Kozgar Mutation Breeding in Chickpea: Perspectives and Prospects for Food Security M. Imran Kozgar Mutation Breeding in Chickpea: Perspectives and Prospects for Food Security Managing Editor: Katarzyna Michalczyk Language Editor: Sara Suliman Published by De Gruyter Open Ltd, Warsaw/Berlin This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 license, which means that the text may be used for non-commercial purposes, provided credit is given to the author. For details go to http://creativecommons.org/licenses/by-nc-nd/3.0/. Copyright © 2014 M. Imran Kozgar ISBN: 978-83-7656-070-0 e-ISBN: 978-83-7656-071-7 Managing Editor: Katarzyna Michalczyk Language Editor: Sara Suliman www.degruyteropen.com Cover illustration: © M. Imran Kozgar Complimentary copy, not for sale. Preface The universal truth is that the agricultural practices and plant breeding strategies are essential tools to feed the inhabitants of the world. These practices become more important in the conditions where population grows rapidly especially the countries like India. In order to feed the people, the plant breeding strategies has to be developed as per need of an hour and mutation breeding has shown some light to curb the two jargon problems presently persisting in the world vis-à-vis food insecurity and malnutrition. Enhancing the genetic diversity is the prerequisite for utilizing the benefits of successful plant breeding in order to get the necessary yield plateau and induced mutagenesis is one of the techniques which have potential -

Tc Akdeniz Üniversitesi Fen Bilimleri Enstitüsü Türkiye
T.C. AKDENİZ ÜNİVERSİTESİ FEN BİLİMLERİ ENSTİTÜSÜ TÜRKİYE’DE ENDEMİK OLARAK YETİŞEN Cicer isauricum P.H. DAVİS’İN YAYILIŞI VE CANLI STRESLERİ Mehmet TEKİN YÜKSEK LİSANS TEZİ TARLA BİTKİLERİ ANABİLİM DALI 2016 ÖZET TÜRKİYE’DE ENDEMİK OLARAK YETİŞEN Cicer isauricum P.H. DAVİS’İN YAYILIŞI VE CANLI STRESLERİ Mehmet TEKİN Yüksek Lisans Tezi, Tarla Bitkileri Anabilim Dalı Danışman: Prof. Dr. Cengiz TOKER Mayıs 2016, 61 Sayfa Fabaceae familyası içerisinde bulunan Cicer L. cinsi 49 taksona sahiptir ve bu taksonlardan 17’si ülkemizde yayılış göstermektedir. Bu çalışma ile Türkiye’de endemik olarak yetişen çok yıllık nohut türlerinden biri olan Cicer isauricum P.H. Davis’in yayılış alanları ve canlı stres etmenleri belirlenmiştir. Endemik olan bu türün sadece ülkemizin C3 karesinin doğusunda (Akseki/Antalya, Alanya/Antalya, Hadim/Konya ve çevresinde) yayılış gösterdiği bilinmekteydi. Ancak bu çalışma ile Antalya kent merkezini kuzeybatıdan çevreleyen Güllük dağlarında bulunan Geyikbayırı köyünde bu türe ait yeni populasyonlar keşfedilmiştir. Türün C3 karesinin genelinde potansiyel yayılım alanı yaklaşık olarak 7000 km2’dir. Bu verilere göre; IUCN (Dünya Doğa ve Doğal Kaynakları Koruma Birliği) tehlike kategorisi “EN” (tehlikede) olarak güncellenmiştir. Yapılan incelemeler sonucunda tür üzerinde etkisi olan canlı stres etmenleri belirlenmiştir. Birincisi nohut bitkisinin en büyük zararlılarından olan yaprak ve taze baklaları tüketerek yaşamını sürdüren yeşil kurt (Helicoverpa armigera Hübner syn. Heliothis armigera Hübner)’tur. Buna ek olarak, çok yaygın olmasa da tür etrafında parazit olarak yaşayan canavar otu (Orobanche sp.) türleri de tespit edilmiştir. Yeşil kurt ve canavar otu türün yeni populasyonlarının keşfedildiği bölgelerde görülmüş ve fenotipik olarak gözlenmiştir. Diğer bir canlı stresi nohutta (Cicer arietinum L.) verim kaybına yol açan en önemli hastalık olan nohut yanıklık hastalığı (antraknoz)’dır.