No Association Between ACTN3 R577X and ACE I/D Polymorphisms and Endurance Running Times in 698 Caucasian Athletes Ioannis D
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Runtastic Running Season Is Here Final1000px
RUNNING SEASON IS HERE! Temperatures are rising and your activity level should be, too! The good news? There are literally millions of races & events worldwide to keep you focused, motivated and feeling supported. Whether you’d like to give your first 5K a go or you’re looking to set a new marathon PR this year – use the engaging facts & figures below to get inspired, learn about fellow runners in the Runtastic Community and consider what personal challenges (races or otherwise) you might like to commit yourself to this running season. Ready… Set… GO! Statistically, men run START Listening to music their fastest while running can boost marathon at age 27; performance by up to women at 29.1 15%. 2 27 29 0 10 20 30 AGE SCALE The fastest half marathon (female) in the world was run by Florence Jebet Kiplagat in 1:05:12.3 The fastest marathon Male runners (male) in the world was run by covering more than Dennis Kipruto Kimetto in 30 miles 2:02:57.3 a week are likely to conceive girls. 4 In 2013, over 42 million Americans ran six or more days per week.5 44.6 million pairs of running shoes were sold in the United States in 2012, totaling $3.04 billion! 6 The London Marathon is the Guinness World Record's largest annual fundraising event in the world. Since its launch, runners have raised more than £500 million for charity! 7 £ £ The average 10K time for Runtastic users is 56 min. 19 sec. for men and 63 min. -

Iaaf World Half Marathon Championships Kavarna 2012 Facts & Figures
IAAF WORLD HALF MARATHON CHAMPIONSHIPS KAVARNA 2012 FACTS & FIGURES Incorporating the IAAF World Half Marathon Championships (1992-2005/2008- 2010) & the IAAF World Road Running Championships 2006/2007 Past Championships...............................................................................................1 Past Medallists .......................................................................................................1 Overall Placing Table..............................................................................................5 Most Medals Won...................................................................................................6 Youngest & Oldest..................................................................................................7 Most Appearances by athlete.................................................................................7 Most Appearances by country................................................................................8 Competitorʼs Index ...............................................................................................10 World Road Race Records & Best Performances ...............................................33 Progression of World Record & Best Performances at Half Marathon ................35 KAVARNA 2012 ★ FACTS & FIGURES/PAST CHAMPS & MEDALLISTS 1 PAST CHAMPIONSHIPS –––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––– These totals have been adjusted from those previously published so that athletes entered for the championships -
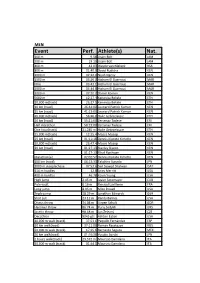
Event Perf. Athlete(S) Nat
MEN Event Perf. Athlete(s) Nat. 100 m 9.58 Usain Bolt JAM 200 m 19.19 Usain Bolt JAM 400 m 43.03 Wayde van Niekerk RSA 800 m 01:40.9 David Rudisha KEN 1000 m 02:12.0 Noah Ngeny KEN 1500 m 03:26.0 Hicham El Guerrouj MAR Mile 03:43.1 Hicham El Guerrouj MAR 2000 m 04:44.8 Hicham El Guerrouj MAR 3000 m 07:20.7 Daniel Komen KEN 5000 m 12:37.4 Kenenisa Bekele ETH 10,000 m(track) 26:17.5 Kenenisa Bekele ETH 10 km (road) 26:44:00 Leonard Patrick Komon KEN 15 km (road) 41:13:00 Leonard Patrick Komon KEN 20,000 m(track) 56:26.0 Haile Gebrselassie ETH 20 km (road) 55:21:00 Zersenay Tadese ERI Half marathon 58:23:00 Zersenay Tadese ERI One hour(track) 21,285 m Haile Gebrselassie ETH 25,000 m(track) 12:25.4 Moses Mosop KEN 25 km (road) 01:11:18 Dennis Kipruto Kimetto KEN 30,000 m(track) 26:47.4 Moses Mosop KEN 30 km (road) 01:27:13 Stanley Biwott KEN 01:27:13 Eliud Kipchoge KEN Marathon[a] 02:02:57 Dennis Kipruto Kimetto KEN 100 km (road) 06:13:33 Takahiro Sunada JPN 3000 m steeplechase 07:53.6 Saif Saaeed Shaheen QAT 110 m hurdles 12.8 Aries Merritt USA 400 m hurdles 46.78 Kevin Young USA High jump 2.45 m Javier Sotomayor CUB Pole vault 6.16 m Renaud Lavillenie FRA Long jump 8.95 m Mike Powell USA Triple jump 18.29 m Jonathan Edwards GBR Shot put 23.12 m Randy Barnes USA Discus throw 74.08 m Jürgen Schult GDR Hammer throw 86.74 m Yuriy Sedykh URS Javelin throw 98.48 m Jan Železný CZE Decathlon 9045 pts Ashton Eaton USA 10,000 m walk (track) 37:53.1 Paquillo Fernández ESP 10 km walk(road) 37:11:00 Roman Rasskazov RUS 20,000 m walk (track) 17:25.6 Bernardo -

START LIST Marathon Men - Final
London World Championships 4-13 August 2017 START LIST Marathon Men - Final RECORDS RESULT NAME COUNTRY AGE VENUE DATE World Record WR 2:02:57 Dennis Kipruto KIMETTO KEN 30 Berlin 28 Sep 2014 Championships Record CR 2:06:54 Abel KIRUI KEN 27 Berlin 22 Aug 2009 World Leading WL 2:03:58 Wilson Kipsang KIPROTICH KEN 35 Tokyo 26 Feb 2017 6 August 2017 10:55 START TIME NAME COUNTRY DATE of BIRTH PERSONAL BEST SEASON BEST Mariano MASTROMARINO ARG 15 Sep 82 2:15:27 Jack COLREAVY AUS 15 Jun 89 2:18:32 2:18:32 Josh HARRIS AUS 3 Jul 90 2:17:08 2:17:08 Brad MILOSEVIC AUS 26 May 89 2:16:00 2:16:24 Valentin PFEIL AUT 17 Jul 88 2:14:50 2:14:50 Abraham NIYONKURU BDI 29 Dec 89 2:16:14 2:16:14 Abdelhadi EL HACHIMI BEL 15 Dec 74 2:10:35 2:26:23 Hassan CHANI BRN 5 May 88 2:14:58 2:14:58 Shumi DECHASA BRN 28 May 89 2:06:43 2:15:35 Eric GILLIS CAN 8 Mar 80 2:11:21 Thomas TOTH CAN 6 Oct 91 2:18:58 2:18:58 Manuel CABRERA CHI 30 Jan 83 2:18:04 2:20:19 Leslie ENCINA CHI 2 Jan 82 2:17:46 2:20:59 Enzo YÁÑEZ CHI 13 Sep 85 2:17:35 2:18:39 Abdi Hakin ULAD DEN 14 Jun 91 2:14:03 Mumin GALA DJI 6 Sep 86 2:13:04 Segundo JAMI ECU 12 May 86 2:17:11 2:18:04 Yohanes GHEBREGERGIS ERI 1 Jan 89 2:08:14 2:08:14 Ghebrezgiabhier KIBROM ERI 1 Feb 87 2:09:36 2:14:52 Amanuel MESEL ERI 29 Dec 90 2:08:17 2:10:44 Iván FERNÁNDEZ ESP 10 Jun 88 2:12:55 Javier GUERRA ESP 10 Nov 83 2:09:33 2:10:55 Ayad LAMDASSEM ESP 11 Oct 81 2:09:28 2:12:30 Roman FOSTI EST 6 Jun 83 2:17:54 2:28:20 Tiidrek NURME EST 18 Nov 85 2:17:59 Tsegaye MEKONNEN ETH 15 Jun 95 2:04:32 2:07:26 Tamirat TOLA ETH 11 Aug 91 -

Cardiff 2016 F&F
IAAF/Cardiff University WORLD HALF MARATHON CHAMPIONSHIPS FACTS & FIGURES Incorporating the IAAF World Half Marathon Championships (1992-2005/2008-2010-2012-2014) IAAF World Road Running Championships 2006/2007 Past Championships...............................................................................................1 Past Medallists .......................................................................................................1 Overall Placing Table..............................................................................................5 Most Medals Won...................................................................................................6 Youngest & Oldest..................................................................................................7 Most Appearances by athlete.................................................................................7 Most Appearances by country................................................................................8 Country Index .......................................................................................................10 World Road Race Records & Best Performances at 15Km, 20Km & Half Mar .........39 Progression of World Record & Best Performance at 15Km, 20Km & Half Mar .......41 CARDIFF 2016 ★ FACTS & FIGURES/PAST CHAMPS & MEDALLISTS 1 PAST CHAMPIONSHIPS –––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––– These totals do not include athletes entered for the championships who never started, but teams are counted -
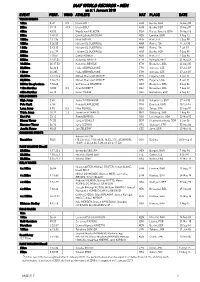
IAAF WORLD RECORDS - MEN As at 1 January 2018 EVENT PERF
IAAF WORLD RECORDS - MEN as at 1 January 2018 EVENT PERF. WIND ATHLETE NAT PLACE DATE TRACK EVENTS 100m 9.58 0.9 Usain BOLT JAM Berlin, GER 16 Aug 09 200m 19.19 -0.3 Usain BOLT JAM Berlin, GER 20 Aug 09 400m 43.03 Wayde van NIEKERK RSA Rio de Janeiro, BRA 14 Aug 16 800m 1:40.91 David Lekuta RUDISHA KEN London, GBR 9 Aug 12 1000m 2:11.96 Noah NGENY KEN Rieti, ITA 5 Sep 99 1500m 3:26.00 Hicham EL GUERROUJ MAR Roma, ITA 14 Jul 98 1 Mile 3:43.13 Hicham EL GUERROUJ MAR Roma, ITA 7 Jul 99 2000m 4:44.79 Hicham EL GUERROUJ MAR Berlin, GER 7 Sep 99 3000m 7:20.67 Daniel KOMEN KEN Rieti, ITA 1 Sep 96 5000m 12:37.35 Kenenisa BEKELE ETH Hengelo, NED 31 May 04 10,000m 26:17.53 Kenenisa BEKELE ETH Bruxelles, BEL 26 Aug 05 20,000m 56:26.0 Haile GEBRSELASSIE ETH Ostrava, CZE 27 Jun 07 1 Hour 21,285 Haile GEBRSELASSIE ETH Ostrava, CZE 27 Jun 07 25,000m 1:12:25.4 Moses Cheruiyot MOSOP KEN Eugene, USA 3 Jun 11 30,000m 1:26:47.4 Moses Cheruiyot MOSOP KEN Eugene, USA 3 Jun 11 3000m Steeplechase 7:53.63 Saif Saaeed SHAHEEN QAT Bruxelles, BEL 3 Sep 04 110m Hurdles 12.80 0.3 Aries MERRITT USA Bruxelles, BEL 7 Sep 12 400m Hurdles 46.78 Kevin YOUNG USA Barcelona, ESP 6 Aug 92 FIELD EVENTS High Jump 2.45 Javier SOTOMAYOR CUB Salamanca, ESP 27 Jul 93 Pole Vault 6.16i Renaud LAVILLENIE FRA Donetsk, UKR 15 Feb 14 Long Jump 8.95 0.3 Mike POWELL USA Tokyo, JPN 30 Aug 91 Triple Jump 18.29 1.3 Jonathan EDWARDS GBR Göteborg, SWE 7 Aug 95 Shot Put 23.12 Randy BARNES USA Los Angeles, USA 20 May 90 Discus Throw 74.08 Jürgen SCHULT GDR Neubrandenburg, GDR 6 Jun 86 Hammer Throw -

Revista Corricolari Es + Correr Digital + Tarjeta € *Tarjeta Con Acceso a Ventajas 36 /Año Revista Papel + Tarjeta Vlrd N20,90 En *Valorado
CO-327-084.084 PUBLI MAPOMA.qxp:Maquetación 1 28/01/15 20:41 Página 84 CO-327-003.003 SUMARIO.qxp:Maquetación 1 29/01/15 17:15 Página 3 número 327 / febrero 2015 / www.corricolari.es CONTENIDOS EL ENIGMA AFRICANO: ¿POR QUÉ GANAN TODAS LAS COMPETICIONES? PÁG.14 Correr con sueños en la cabeza “Un atleta no puede correr con dinero en los APRENDE A BUSCAR LAS FRONTERAS DE TU CUERPO: bolsillos. Debe hacerlo con fe en su corazón y SOBREENTRENAMIENTO sueños en su cabeza”. Esta bonita cita pertenece a PÁG. 34 Emil Zatopek, atleta checoslovaco que alcanzó la • Homenaje a Emilio Ponce / PAG. 4 cima del fondo mundial a finales de los años pone en juego cuarenta y principios de los cincuenta. El mensaje •Londres que quiere transmitir es sencillo: hay que correr el récord del mundo de maratón / PAG. 6 para vivir, realizar un camino en búsqueda de la • Julio Albornos, el hombre de las medias maratones / PAG. 10 felicidad, que está muchas veces al final de nuestras • Diario de un maratoniano. Entrenando a alto nivel / PAG. 12 zancadas. Hay muchos atletas que, como predica • El enigma africano: ¿por qué lo ganan todo? / Zatopek, corren con sueños en su cabeza. PAG. 14 Completar un maratón, llegar a la meta de una • Entrenamiento en cuestas: carrera con tus hijos de la mano, acabar tu primera así debe ser la distancia e inclinación / PAG. 18 prueba de diez kilómetros o apuntarte a una carrera •Clavescotidianas para adelgazar: de montaña son desafíos con los que muchas no basta solo con correr / PAG. -

Cardiff 2016 F&F.Qxp WHM F&F
IAAF/Cardiff University WORLD HALF MARATHON CHAMPIONSHIPS FACTS & FIGURES Incorporating the IAAF World Half Marathon Championships (1992-2005/2008-2010-2012-2014) IAAF World Road Running Championships 2006/2007 Past Championships...............................................................................................1 Past Medallists .......................................................................................................1 Overall Placing Table..............................................................................................5 Most Medals Won...................................................................................................6 Youngest & Oldest..................................................................................................7 Most Appearances by athlete.................................................................................7 Most Appearances by country................................................................................8 Country Index .......................................................................................................10 World Road Race Records & Best Performances at 15Km, 20Km & Half Mar .........39 Progression of World Record & Best Performance at 15Km, 20Km & Half Mar .......41 CARDIFF 2016 ★ FACTS & FIGURES/PAST CHAMPS & MEDALLISTS 1 PAST CHAMPIONSHIPS –––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––– These totals do not include athletes entered for the championships who never started, but teams are counted -

Auf Der Suche Nach Dem Heiligen Gral
30 Nummer 64 Freitag, 17. März 2017 Sport Sport am Wochenende Fall Gabius: FUSSBALL Förderung nur bei Dritte Liga: SV Werder Bremen II - SC Preußen Münster (Fr Auf der Suche nach 19), FSV Zwickau - FC Hansa Rostock, MSV Duisburg - FC Rot- Weiß Erfurt, VfR Aalen - FSV Frankfurt, VfL Osnabrück - Start im DLV-Trikot Chemnitzer FC, SC Paderborn - SSV Jahn Regensburg, SC Fortuna Köln - SG Großaspach, FSV Mainz II - Holstein Kiel (alle Sa 14), VfL Sportfreunde Lotte - Hallescher FC, FC Fehlende Wertschätzung? Verband Magdeburg - SV Wehen Wiesbaden (beide So 14). dem Heiligen Gral kontert Kritik des Marathonläufers Regionalliga: TSG 1899 Hoffenheim II - TSV Steinbach, 1. FC Kaiserslautern II - SSV Ulm, FC Homburg - KSV Hessen Kassel, SV Elversberg - TuS Koblenz (alle Fr 19), SV Eintr. Trier Mit Hochdruck wird daran gearbeitet, einen Marathon unter zwei Stunden zu laufen STUTTGART (dpa). Wer nicht im Deutsch- - Wormatia Worms, SC Teutonia Watzenborn-Steinberg - FK landtrikot startet, wird vom Verband auch Pirmasens, Stuttgarter Kickers - VfB Stuttgart II, FC Nöttingen - 1. FC Saarbrücken (alle Sa 14), SV Waldhof nicht finanziell gefördert. Dieses Grund- Mannheim - Kickers Offenbach (So 14). prinzip betonte DLV-Chef Clemens Prokop, Oberliga: SC Freiburg II - SV Sandhausen II, Göppinger SV - als er die Kritik von Marathon-Rekordhalter Offenburger FV (beide Sa 14), SSV Reutlingen - SpVgg Arne Gabius am Deutschen Leichtathletik- Neckarelz, Neckarsulmer SU - FV Ravensburg, Spielberg- FSV Bissingen, Karlsruher SC II - FC Ast. Walldorf II, CfR Verband konterte. „Wir haben ihm 2016 Pforzheim - Stuttgarter Kickers II, FSV Hollenbach - SV sämtliche Trainingslager bezahlt“, erklärte Oberachern, TSG Balingen - Bahlinger SC (alle Sa 15). Prokop, „wenn dann seine Planung bei- Verbandsliga: SF Schwäbisch Hall - VfB Neckarrems (Sa spielsweise auf den Berlin-Marathon ab- 14), SGV Freiberg - 1. -

IAAF World Athletics Journalist Award – 2012
IAAF World Athletics Journalist Award – 2012 Elias MAKORI Nationality - Kenyan Born December 26, 1970 Journalistic Career 1991: Diploma in Journalism (NSTI – Now St Augustine University of Tanzania), 1992-1993: The East African Standard – Sports Correspondent 1993-1999: Nation Media Group – Sports Writer 1999 to 2004: Nation Media Group – Deputy Sports Editor, Daily Nation 2004 to date: Nation Media Group – Sports Editor, Daily Nation Elias Makori covered a wide range of top international sporting events from 1994 to date, including the 6th All Africa Games (Harare, Zimbabwe, 1995); Volleyball World Cup (Japan, 1995); football World Cup (2010) and various national preparation camps (1996-1998); World Youth Games (Moscow, 1998); World Cup Cricket (1999 and 2003); Euro 2000 Football Championships and African Nations Cup Football Championships. To fill the void in Kenyan and African athletics journalism, Elias Makori has dedicated his career to track and field, especially since 1999, despite being the Sports Editor at the Daily Nation, one of Africa’s leading English dailies. Has covered several athletics competitions, including the IAAF World Half Marathon Championships, the Beijing and London Olympic Games, the World Cross Country Championships and the World Championships in Athletics. Elias Makori has also worked for and at several top marathons including the Nairobi Marathon, Rotterdam Marathon, Frankfurt Marathon, London Marathon and Amsterdam Marathon. Press Associations: 1995-1999: Vice President – Africa Sports Journalists Union -

MATTONI České Budějovice Half Marathon 8 June 2013 PRESS GUIDE Information for Journalists
MATTONI České Budějovice Half Marathon 8 June 2013 PRESS GUIDE Information for journalists The race is a part of the RunCzech Running League EN www.runczech.com PRESS GUIDE 2 3 PRESS GUIDE Dear sports friends Welcome to České Budějovice! To a city whose history spans over almost seven hundred and fifty years, a Royal city also famed for its mining, minting and university tradition. And as of last year a city boasting a world-class running race. The interest demonstrated by runners and spectators alike in the premiere of the Mattoni České Budějovice Half Marathon was huge, while nobody could have predicted the sensational time scooped up by its winner. Praise is especially due in this respect to Carlo Capalbo and his colleagues who managed to turn Budějovice into a run- ning destination which can now be compared to traditional events organised in other large cities such as those taking place in Berlin and New York. I would like to take this opportunity to wish all those taking part in our second ever half marathon a wonderful time in our city. And in the spirit of our national hero, Emil Zátopek, I’d like to add: “If you want to win something, run 100 meters. If you want to experience something, run the marathon.” Juraj Thoma Mayor of the City of České Budějovice Dear fans of sport and distance running in particular, It’s a pleasure for me to welcome you to the 2nd ever Mattoni České Budějovice Half Marathon. Whether your goal in taking part is about achieving the best possible time, comparing your time against the world running elite, joining a relay with friends or work colleagues, or perhaps joining a race suitable for the whole family, including your children, we’re delighted to see you here. -

MATTONI Olomouc Half Marathon 22 June 2013 PRESS GUIDE Information for Journalists
MATTONI Olomouc Half Marathon 22 June 2013 PRESS GUIDE Information for journalists The race is a part of the RunCzech Running League EN www.runczech.com PRESS GUIDE 2 3 PRESS GUIDE This year Olomouc will welcome runners to the city for the fourth time already! On 22nd June, crowds of eager participants will set off on the course inscribing one more Mattoni Half Marathon Olomouc on the city’s sporting history forever. The first chapter of the running series in Olomouc got underway in 2010. Even back then, the race managed to entice 1,600 runners from 23 countries around the world. Since that time, this athletics event has made significant headway. Already now firmly established on the RunCzech Running League calendar, the race has evolved into a prestigious event, earning itself fans as well as international acclaim over a very short timeframe. Just as has been the case in the past, this year’s running festival is sure to be a huge success, though of course there will be some new features too. In addition to seeing capacity burgeon to 5,000 eager runners, the race will also bear the unique Silver Quality Label award from the International Association of Athletics Federations (IAAF). This places our city alongside other prominent half marathons boasting the award such including those in Yokohama, Rotterdam and Turin. As one of the most beautiful cities in Central Europe, it also makes Olomouc the smallest city in the world to acquire such a high rating. It will be interesting to follow how, together with the organisers, we manage to fulfil the ambitious goals we have set ourselves in light of this award.