Probabilistic Joint Face-Skull Modelling for Facial Reconstruction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Series 1100TDM Tandem MEGALUG Mechanical Joint Restraint
Series 1100TDM Tandem MEGALUG® Mechanical Joint Restraint High Pressure Restraint for Ductile Iron Pipe Features and Applications: • For use on Ductile Iron Pipe 4 inch through 54 inch • High Pressure Restraint • Torque Limiting Twist-Off Nuts • Mechanical Joint follower gland incorporated into the restraint • MEGA-BOND® Coating System For more information on MEGA- BOND, visit our web site at www. ebaa.com • Minimum 2 to 1 Safety Factor Series 1112TDM restraining a mechanical joint fitting. • Constructed of A536 Ductile Iron Post Pressure Rating • EBAA-Seal™ Mechanical Nominal Pipe Shipping Assembly (PSI) Joint Gaskets are provided Size Weights* Deflection with all 1100TDM MEGALUG 4 21.6 3° 700 restraints. These are required 6 33.0 3° 700 to accommodate the pressure ratings and safety factors 8 40.0 3° 700 shown. 10 60.2 3° 700 12 75.0 3° 700 • New: High strength heavy hex 14 112.7 2° 700 machine bolts with T-nuts are 16 131.6 2° 700 provided to facilitate easier assembly due to the fittings 18 145.2 1½° 500 radius area prohibiting the use 20 166.6 1½° 500 longer T-bolts. 24 290.2 1½° 500 30 457.9 1° 500 • T-Nuts constructed of High 36 553.63 1° 500 Tensile Ductile Iron with Fluropolymer Coating. 42 1,074.8 1° 500 48 1,283.1 1° 500 For use on water or wastewater 54 1,445.32 ½° 400 pipelines subject to hydrostatic NOTE: For applications or pressures other than those shown please pressure and tested in accordance contact EBAA for assistance. -

Chapter 14. Anthropometry and Biomechanics
Table of contents 14 Anthropometry and biomechanics........................................................................................ 14-1 14.1 General application of anthropometric and biomechanic data .....................................14-2 14.1.1 User population......................................................................................................14-2 14.1.2 Using design limits ................................................................................................14-4 14.1.3 Avoiding pitfalls in applying anthropometric data ................................................14-6 14.1.4 Solving a complex sequence of design problems ..................................................14-7 14.1.5 Use of distribution and correlation data...............................................................14-11 14.2 Anthropometric variability factors..............................................................................14-13 14.3 Anthropometric and biomechanics data......................................................................14-13 14.3.1 Data usage............................................................................................................14-13 14.3.2 Static body characteristics....................................................................................14-14 14.3.3 Dynamic (mobile) body characteristics ...............................................................14-28 14.3.3.1 Range of whole body motion........................................................................14-28 -

Medical Terminology Abbreviations Medical Terminology Abbreviations
34 MEDICAL TERMINOLOGY ABBREVIATIONS MEDICAL TERMINOLOGY ABBREVIATIONS The following list contains some of the most common abbreviations found in medical records. Please note that in medical terminology, the capitalization of letters bears significance as to the meaning of certain terms, and is often used to distinguish terms with similar acronyms. @—at A & P—anatomy and physiology ab—abortion abd—abdominal ABG—arterial blood gas a.c.—before meals ac & cl—acetest and clinitest ACLS—advanced cardiac life support AD—right ear ADL—activities of daily living ad lib—as desired adm—admission afeb—afebrile, no fever AFB—acid-fast bacillus AKA—above the knee alb—albumin alt dieb—alternate days (every other day) am—morning AMA—against medical advice amal—amalgam amb—ambulate, walk AMI—acute myocardial infarction amt—amount ANS—automatic nervous system ant—anterior AOx3—alert and oriented to person, time, and place Ap—apical AP—apical pulse approx—approximately aq—aqueous ARDS—acute respiratory distress syndrome AS—left ear ASA—aspirin asap (ASAP)—as soon as possible as tol—as tolerated ATD—admission, transfer, discharge AU—both ears Ax—axillary BE—barium enema bid—twice a day bil, bilateral—both sides BK—below knee BKA—below the knee amputation bl—blood bl wk—blood work BLS—basic life support BM—bowel movement BOW—bag of waters B/P—blood pressure bpm—beats per minute BR—bed rest MEDICAL TERMINOLOGY ABBREVIATIONS 35 BRP—bathroom privileges BS—breath sounds BSI—body substance isolation BSO—bilateral salpingo-oophorectomy BUN—blood, urea, nitrogen -

About Your Knee
OrthoInfo Basics About Your Knee What are the parts of the knee? Your knee is Your knee is made up of four main things: bones, cartilage, ligaments, the largest joint and tendons. in your body Bones. Three bones meet to form your knee joint: your thighbone and one of the (femur), shinbone (tibia), and kneecap (patella). Your patella sits in most complex. front of the joint and provides some protection. It is also vital Articular cartilage. The ends of your thighbone and shinbone are covered with articular cartilage. This slippery substance to movement. helps your knee bones glide smoothly across each other as you bend or straighten your leg. Because you use it so Two wedge-shaped pieces of meniscal cartilage act as much, it is vulnerable to Meniscus. “shock absorbers” between your thighbone and shinbone. Different injury. Because it is made from articular cartilage, the meniscus is tough and rubbery to help up of so many parts, cushion and stabilize the joint. When people talk about torn cartilage many different things in the knee, they are usually referring to torn meniscus. can go wrong. Knee pain or injury Femur is one of the most (thighbone) common reasons people Patella (kneecap) see their doctors. Most knee problems can be prevented or treated with simple measures, such as exercise or Articular cartilage training programs. Other problems require surgery Meniscus to correct. Tibia (shinbone) 1 OrthoInfo Basics — About Your Knee What are ligaments and tendons? Ligaments and tendons connect your thighbone Collateral ligaments. These are found on to the bones in your lower leg. -

Study Guide Medical Terminology by Thea Liza Batan About the Author
Study Guide Medical Terminology By Thea Liza Batan About the Author Thea Liza Batan earned a Master of Science in Nursing Administration in 2007 from Xavier University in Cincinnati, Ohio. She has worked as a staff nurse, nurse instructor, and level department head. She currently works as a simulation coordinator and a free- lance writer specializing in nursing and healthcare. All terms mentioned in this text that are known to be trademarks or service marks have been appropriately capitalized. Use of a term in this text shouldn’t be regarded as affecting the validity of any trademark or service mark. Copyright © 2017 by Penn Foster, Inc. All rights reserved. No part of the material protected by this copyright may be reproduced or utilized in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system, without permission in writing from the copyright owner. Requests for permission to make copies of any part of the work should be mailed to Copyright Permissions, Penn Foster, 925 Oak Street, Scranton, Pennsylvania 18515. Printed in the United States of America CONTENTS INSTRUCTIONS 1 READING ASSIGNMENTS 3 LESSON 1: THE FUNDAMENTALS OF MEDICAL TERMINOLOGY 5 LESSON 2: DIAGNOSIS, INTERVENTION, AND HUMAN BODY TERMS 28 LESSON 3: MUSCULOSKELETAL, CIRCULATORY, AND RESPIRATORY SYSTEM TERMS 44 LESSON 4: DIGESTIVE, URINARY, AND REPRODUCTIVE SYSTEM TERMS 69 LESSON 5: INTEGUMENTARY, NERVOUS, AND ENDOCRINE S YSTEM TERMS 96 SELF-CHECK ANSWERS 134 © PENN FOSTER, INC. 2017 MEDICAL TERMINOLOGY PAGE III Contents INSTRUCTIONS INTRODUCTION Welcome to your course on medical terminology. You’re taking this course because you’re most likely interested in pursuing a health and science career, which entails proficiencyincommunicatingwithhealthcareprofessionalssuchasphysicians,nurses, or dentists. -

Monitoring Methods of Human Body Joints: State-Of-The-Art and Research Challenges
sensors Review Monitoring Methods of Human Body Joints: State-of-the-Art and Research Challenges Abu Ilius Faisal 1, Sumit Majumder 1 , Tapas Mondal 2, David Cowan 3, Sasan Naseh 1 and M. Jamal Deen 1,* 1 Department of Electrical and Computer Engineering, McMaster University, Hamilton, ON L8S 4L8, Canada; [email protected] (A.I.F.); [email protected] (S.M.); [email protected] (S.N.) 2 Department of Pediatrics, McMaster University, Hamilton, ON L8S 4L8, Canada; [email protected] 3 Department of Medicine, St. Joseph’s Healthcare Hamilton, Hamilton, ON L8N 4A6, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-905-5259-140 (ext. 27137) Received: 26 April 2019; Accepted: 4 June 2019; Published: 10 June 2019 Abstract: The world’s population is aging: the expansion of the older adult population with multiple physical and health issues is now a huge socio-economic concern worldwide. Among these issues, the loss of mobility among older adults due to musculoskeletal disorders is especially serious as it has severe social, mental and physical consequences. Human body joint monitoring and early diagnosis of these disorders will be a strong and effective solution to this problem. A smart joint monitoring system can identify and record important musculoskeletal-related parameters. Such devices can be utilized for continuous monitoring of joint movements during the normal daily activities of older adults and the healing process of joints (hips, knees or ankles) during the post-surgery period. A viable monitoring system can be developed by combining miniaturized, durable, low-cost and compact sensors with the advanced communication technologies and data processing techniques. -

Sticky Lips BBQ Virtual Tour Sticky Lips BBQ Point Your Phone at the Juke Joint QR Codes Below
33 Taps! full bar! MUSEUM & BEER EMPORIUM Texas Style Dining Dine-in Counter Ordering or Full Service Slow Smokin’ the Best Grilled and Pit Styles from the Legendary Barbeque Regions Across America. Sticky Lips BBQ Virtual Tour Sticky Lips BBQ Point your phone at the Juke Joint QR Codes below. (585) 292-5544 830 Jefferson Road Henrietta, NY 14623 www.StickyLipsBBQ.com Take a virtual walk around the restaurant, courtesy of Virtual Space Productions! Visit our kissin’ cousin Sticky Soul & BBQ (585) 288-1910 625 Culver Rd. Rochester, 14609 www.StickySoulAndBBQ.com Experience the BLUES AND BBQ TOUR as a music video! 2021v17 French Fried Basket Home-Cut Style Fries.................................................... 5.49 Loaded with Chili, Cheddar Jack Cheese, Lip Smackers Onions, & Pickled Jalapenos................................ 9.49 Sweet Potato Fries......................................................... 5.49 Deep Fried Pickles NEW GIGANTIC Chicken Wings Kosher dill pickle spears, battered and breaded. Served with our spicy Cajun Buttermilk Ranch All orders served with SEASONED POTATO CHIPS. Dressing............................................................................ 8.29 Add THICK N’ CHUNKY BLUE CHEESE... 75¢ Small order... 8.49 Large... 14.99 Extra Large... 28.99 Mississippi Catfish Strips FRIED & GRILLED BBQ Fried then char-grilled. A Brother Wease favorite! Hand-cut catsh strips, Topped with our house-made BBQ sauce. battered and breaded in our special blend of our, and southern style spices. Served with our tangy BUFFALO STYLE Lightly seasoned, fried, and tossed Cajun tartar sauce. in wing sauce. Small (3 pieces)............................................... 8.39 Choose: BUFFALO SAUCE (medium) or SWEET & SOUR. Large (5 pieces)............................................ 12.49 Make it a meal! Add a side for 2.50 GARLIC PARMESAN Lightly seasoned, fried, and tossed in our garlic parmesan sauce. -

Human Anatomy and Physiology
LECTURE NOTES For Nursing Students Human Anatomy and Physiology Nega Assefa Alemaya University Yosief Tsige Jimma University In collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education 2003 Funded under USAID Cooperative Agreement No. 663-A-00-00-0358-00. Produced in collaboration with the Ethiopia Public Health Training Initiative, The Carter Center, the Ethiopia Ministry of Health, and the Ethiopia Ministry of Education. Important Guidelines for Printing and Photocopying Limited permission is granted free of charge to print or photocopy all pages of this publication for educational, not-for-profit use by health care workers, students or faculty. All copies must retain all author credits and copyright notices included in the original document. Under no circumstances is it permissible to sell or distribute on a commercial basis, or to claim authorship of, copies of material reproduced from this publication. ©2003 by Nega Assefa and Yosief Tsige All rights reserved. Except as expressly provided above, no part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system, without written permission of the author or authors. This material is intended for educational use only by practicing health care workers or students and faculty in a health care field. Human Anatomy and Physiology Preface There is a shortage in Ethiopia of teaching / learning material in the area of anatomy and physicalogy for nurses. The Carter Center EPHTI appreciating the problem and promoted the development of this lecture note that could help both the teachers and students. -

Human Vitamin and Mineral Requirements
Human Vitamin and Mineral Requirements Report of a joint FAO/WHO expert consultation Bangkok, Thailand Food and Agriculture Organization of the United Nations World Health Organization Food and Nutrition Division FAO Rome The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concern- ing the delimitation of its frontiers or boundaries. All rights reserved. Reproduction and dissemination of material in this information product for educational or other non-commercial purposes are authorized without any prior written permission from the copyright holders provided the source is fully acknowledged. Reproduction of material in this information product for resale or other commercial purposes is prohibited without written permission of the copyright holders. Applications for such permission should be addressed to the Chief, Publishing and Multimedia Service, Information Division, FAO, Viale delle Terme di Caracalla, 00100 Rome, Italy or by e-mail to [email protected] © FAO 2001 FAO/WHO expert consultation on human vitamin and mineral requirements iii Foreword he report of this joint FAO/WHO expert consultation on human vitamin and mineral requirements has been long in coming. The consultation was held in Bangkok in TSeptember 1998, and much of the delay in the publication of the report has been due to controversy related to final agreement about the recommendations for some of the micronutrients. A priori one would not anticipate that an evidence based process and a topic such as this is likely to be controversial. -
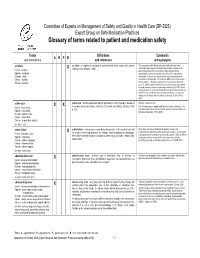
Glossary of Terms Related to Patient and Medication Safety
Committee of Experts on Management of Safety and Quality in Health Care (SP-SQS) Expert Group on Safe Medication Practices Glossary of terms related to patient and medication safety Terms Definitions Comments A R P B and translations and references and synonyms accident accident : an unplanned, unexpected, and undesired event, usually with adverse “For many years safety officials and public health authorities have Xconsequences (Senders, 1994). discouraged use of the word "accident" when it refers to injuries or the French : accident events that produce them. An accident is often understood to be Spanish : accidente unpredictable -a chance occurrence or an "act of God"- and therefore German : Unfall unavoidable. However, most injuries and their precipitating events are Italiano : incidente predictable and preventable. That is why the BMJ has decided to ban the Slovene : nesreča word accident. (…) Purging a common term from our lexicon will not be easy. "Accident" remains entrenched in lay and medical discourse and will no doubt continue to appear in manuscripts submitted to the BMJ. We are asking our editors to be vigilant in detecting and rejecting inappropriate use of the "A" word, and we trust that our readers will keep us on our toes by alerting us to instances when "accidents" slip through.” (Davis & Pless, 2001) active error X X active error : an error associated with the performance of the ‘front-line’ operator of Synonym : sharp-end error French : erreur active a complex system and whose effects are felt almost immediately. (Reason, 1990, This definition has been slightly modified by the Institute of Medicine : “an p.173) error that occurs at the level of the frontline operator and whose effects are Spanish : error activo felt almost immediately.” (Kohn, 2000) German : aktiver Fehler Italiano : errore attivo Slovene : neposredna napaka see also : error active failure active failures : actions or processes during the provision of direct patient care that Since failure is a term not defined in the glossary, its use is not X recommended. -

Cranial Kinesis in Lepidosaurs: Skulls in Motion
Topics in Functional and Ecological Vertebrate Morphology, pp. 15-46. P. Aerts, K. D’Août, A. Herrel & R. Van Damme, Eds. © Shaker Publishing 2002, ISBN 90-423-0204-6 Cranial Kinesis in Lepidosaurs: Skulls in Motion Keith Metzger Department of Anatomical Sciences, Stony Brook University, USA. Abstract This chapter reviews various aspects of cranial kinesis, or the presence of moveable joints within the cranium, with a concentration on lepidosaurs. Previous studies tend to focus on morphological correlates of cranial kinesis, without taking into account experimental evidence supporting or refuting the presence of the various forms of cranial kinesis in these taxa. By reviewing experimental and anatomical evidence, the validity of putative functional hypotheses for cranial kinesis in lepidosaurs is addressed. These data are also considered with respect to phylogeny, as such an approach is potentially revealing regarding the development of various forms of cranial kinesis from an evolutionary perspective. While existing evidence does not allow for events leading to the origin of cranial kinesis in lepidosaurs to be clearly understood at the present time, the potential role of exaptation in its development for specific groups (i.e., cordylids, gekkonids, varanids) is considered here. Directions for further research include greater understanding of the distribution of cranial kinesis in lepidosaurs, investigation of intraspecific variation of this feature (with a focus on ontogenetic factors and prey properties as variables which may influence the presence of kinesis), and continued study of the relationship between experimentally proven observation of cranial kinesis and cranial morphology. Key words: cranial kinesis, lepidosaur skull, functional morphology. Introduction Cranial kinesis, or the presence of moveable joints within the cranium, has been a subject of considerable interest to researchers for more than a century (for references see Bock, 1960; Frazzetta, 1962; Smith, 1982). -

Complete Tear of the Radial Collateral Ligament of the Third Metacarpophalangeal Joint
■ trauma update Complete Tear of the Radial Collateral Ligament of the Third Metacarpophalangeal Joint ALAN E. FREELAND, MD; E. RHETT HOBGOOD, MD plane perpendicular to the head and course distally and long anteroposterior diameter obliquely to insert on the radi- Early repair of complete collateral ligament tears restores of the metacarpal head. The al and ulnar tuberosities on the joint alignment, stability, and full passive range of motion. glenoid cavity of the proximal palmar proximal margins of phalanx covers most of the the base of the proximal pha- metacarpal head articular sur- lanx. The radial collateral liga- he metacarpophalangeal Some rotation and deviation face in the transverse plane, ment originates and attaches Tjoints of the fingers are also occur during flexion and but only approximately one- slightly closer to the joint mar- complex condylar hinge joints with pinching and grasping third of its facade in the sagit- gin and courses less vertically capable of multiplanar motions. tal plane at any point during than its ulnar counterpart. The motion.1-3 The joint surfaces flexion and extension. The accessory collateral ligaments and surrounding capsule and ANATOMY volar plate elongates the gle- originate just palmar to the ligaments provide static stabil- The convex metacarpal ity whereas the intrinsic and head is elliptically shaped and extrinsic muscles are dynamic has a correspondingly variable stabilizers. The normal joint radius of curvature in the mid- Radial and ulnar collateral ligament injuries flexes approximately 90° and sagittal plane, whereas the may extend as much as 20°- base of the proximal phalanx are evenly distributed in the middle finger, 30°.