12 August 2021 Aperto
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Elenco Accessi in in Proprietà Private ANDEZENO
RACCOLTA E GESTIONE DEI RIFIUTI E SERVIZI DI IGIENE URBANA CAPITOLATO SPECIALE D’APPALTO ALLEGATO 12 - Elenco accessi in in proprietà private ANDEZENO ANDEZENO VIALE REGINA ELENA 65 UTENZE NON DOMESTICHE ANDEZENO CORSO VITTORIO EMANUELE 121 SALA DANZE BALDISSERO BALDISSERO STRADA BELLAVISTA 9 -11 - 16 BALDISSERO VIA SUPERGA 47 BALDISSERO VIA CHIERI 91 BALDISSERO VIA CHIERI 36 BALDISSERO VIA CHIERI 66 BALDISSERO VIA CHIERI 19 BALDISSERO VIA CHIERI 34 BALDISSERO VIA CHIERI 37 BALDISSERO VIA ROMA 32 BALDISSERO VIA ROMA 34 BALDISSERO VIA CORDOVA 34 BALDISSERO VIA PAVAROLO 9 BALDISSERO VIA TORINO 35 BALDISSERO VIA TORINO 4 BALDISSERO VIA TORINO 7 BALDISSERO VIA TORINO 9 BALDISSERO VIA TORINO 11 BALDISSERO STRADA VIALE 11 BALDISSERO STRADA VIALE 3 BALDISSERO STRADA VIALE 5 BALDISSERO STRADA VIALE 6 BALDISSERO STRADA VIALE 8 BALDISSERO STRADA TETTI RONCHI UTENZE NON DOMESTICHE BALDISSERO VIA CHIERI 70/5 DITTA PRIVATA BALDISSERO VIA CHIERI 85/1 DITTA PRIVATA BALDISSERO VIA CHIERI 85/3 DITTA PRIVATA CARMAGNOLA CARMAGNOLA VICOLO ABRATE TUTTA CARMAGNOLA VIA ALBA PALAZZI NUOVI CARMAGNOLA VIA ALBERTI 19/C CARMAGNOLA VICOLO ANGONOA TUTTA CARMAGNOLA VICOLO ANGONOVA 5 - 6 - 7 - 8 CARMAGNOLA VIA ARBIA CASCINA ELVA CASCINA CARMAGNOLA VIA BARAVALLE TUTTA CARMAGNOLA VIA BELLINO 16 - 18- 20- 22 CARMAGNOLA VIA BELLINO 35 CARMAGNOLA VIA BIBIANA TUTTA CARMAGNOLA VIA BOCCACCIO TUTTA CARMAGNOLA VIA BORGHETTO 15/A - 15/C CARMAGNOLA VIA BOSIO CARMAGNOLA VIA BRAIDA 2/A - 2/B CARMAGNOLA VICOLO BRICCO TUTTA CARMAGNOLA VIA BRICHERASIO 2 - 4 CARMAGNOLA VICOLO BURIASCO -

CV Dott Caresio
F ORMATO EUROPEO PER IL CURRICULUM V I T A E Il sottoscritto Marino Caresio, nato a Ciriè il 15 dicembre 1963, in relazione all’avviso per l’attribuzione dell’incarico di Direttore di Struttura Complessa nella disciplina Ortopedia e Traumatologia per la SC Ortopedia e Traumatologia del Presidio Ospedaliero di Chivasso, dichiara quanto segue: INFORMAZIONI PERSONALI Nome MARINO CARESIO Indirizzo VIA DE GASPERI n. 16 – CIRIE’ ( TO ), 10073 Telefono 011 / 9222315 – 338.6806334 Fax 011 / 94294565 E-mail [email protected] Nazionalità Italiana Data di nascita 15/12/1963 ESPERIENZA LAVORATIVA • Date (da – a) 7/11/1990 – 31/03/1995 • Nome e indirizzo del datore di 1° Clinica Ortopedica Traumatologica - Ospedale CTO di Torino lavoro Diretta dal Prof. Paolo Gallinaro • Tipo di azienda o settore Azienda Sanitaria Ospedaliera C.T.O. – C.R.F. di Torino • Tipo di impiego Medico frequentatore in qualità di Specializzando in Ortopedia e Traumatologia • Principali mansioni e responsabilità Progressiva acquisizione di funzioni professionali specialistiche in Ortopedia attraverso l’attività di Reparto, Ambulatorio, Pronto Soccorso e Sala Operatoria – Negli ultimi due anni: organizzazione delle attività degli Specializzandi frequentatori della 1° Clinica Ortopedica C.T.O. di Torino • Date (da – a) 2/08/1991 – 2/08/1992 • Nome e indirizzo del datore di 7° GRUPPO ARTIGLIERIA DA CAMPAGNA “ADRIA” - Torino lavoro • Tipo di azienda o settore Esercito Italiano • Tipo di impiego Ufficiale Medico • Principali mansioni e responsabilità Sottotenente medico addetto all’infermeria della Caserma “Morelli di Popolo” di Torino, responsabile medico dei Reparti Operativi durante le esercitazioni e i “Campi di Esercitazione” • Date (da – a) 3/04/1995 – 31/10/2007 • Nome e indirizzo del datore di ASL 8 –Divisione di Ortopedia e Traumatologia P.O. -
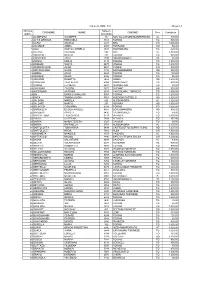
Numero Numero COGNOME NOME COMUNE Prov
Domande AMMESSE Allegato A Numero Numero COGNOME NOME COMUNE Prov. Contributo Ordine Domanda 1 ACAMPORA GIUSEPPE 94 SAN SALVATORE MONFERRATO AL 800,00 2 ACTIS OREGLIA EMANUELA 1348 TORINO TO 900,00 3 AGRO' ANGELO 2828 BRA CN 1.600,00 4 ALI OMAR AMINA 8094 FOSSANO CN 50,00 5 ALOI SANTO CARMELO 5691 MONTANARO TO 520,00 6 ALTANA CLAUDIO 599 ASTI AT 1.800,00 7 ANDREOLI MARZIA 232 TORINO TO 900,00 8 ANDRESINI ANNA 2355 DOMODOSSOLA VB 98,00 9 ANGELI EMILIO 6138 TORINO TO 1.800,00 10 ANSELMI SANTA 4477 VINOVO TO 1.600,00 11 APARASCHIVEI ALINA 5401 CUNEO CN 900,00 12 ARCIDIACONO GILDA 3186 BORGOMANERO NO 900,00 13 ARENA ANNA 5229 TORINO TO 110,00 14 ARGOUB SAAID 8306 TORINO TO 55,00 15 ARMANDI ROSETTA 7459 MOMO NO 1.600,00 16 ARMEANU IULIA OTILIA 5984 MONCALIERI TO 900,00 17 ARORO EZZOHRA 6403 BUSSOLENO TO 55,00 18 ARRIGONI FAUSTO 1873 GHEMME NO 900,00 19 AVERSANO ANTONIO 2833 CASTIGLIONE TORINESE TO 1.600,00 20 BAI ENRICO GIANLUIGI 5907 TORINO TO 1.600,00 21 BAITA SIMONA LUIGIA 7561 OLEGGIO CASTELLO NO 1.600,00 22 BALDONI MIRELLA 836 ALESSANDRIA AL 1.200,00 23 BALLARE' MARCO 251 NOVARA NO 1.600,00 24 BALOCCO GIOVANNA 6108 TORINO TO 1.600,00 25 BARBAGLIA ELISEO ANGELO 4076 BORGOMANERO NO 900,00 26 BARBIERI RIPALTA 8416 CARMAGNOLA TO 89,00 27 BARCELLONA FRANCESCO 5117 VENARIA TO 1.600,00 28 BARRA CATERINA 7854 VENARIA TO 467,00 29 BARRA MARIA TERESA 6325 CAVOUR TO 90,00 30 BARRA RAFFAELLA 6761 ALESSANDRIA AL 2.100,00 31 BARTOLOTTA TOMMASINA 5494 CASTELLETTO SOPRA TICINO NO 1.200,00 32 BARTOLUCCI NADIA 7682 ALBA CN 1.600,00 33 BASHMETA MAKBULE -

Economic Inequality in Northwestern Italy: a Long-Term View (Fourteenth to Eighteenth Centuries)
Guido Alfani PAM - Bocconi University Dondena Centre and IGIER Via Roentgen 1, 20136 Milan - Italy [email protected] Economic inequality in northwestern Italy: a long-term view (fourteenth to eighteenth centuries) (provisional version – please ask for author’s consent to quote) 1 Abstract This article provides a comprehensive picture of economic inequality in northwestern Italy (Piedmont), focusing on the long-term developments occurred during 1300-1800 ca. Regional studies of this kind are rare, and none of them has as long a timescale. The new data proposed illuminate many little-known aspects of wealth distribution and general economic inequality in preindustrial times, and support the idea that during the Early Modern period, inequality grew everywhere: both in cities and in rural areas, and independently from whether the economy was growing or stagnating. This finding challenges earlier views that explained inequality growth as the consequence of economic development. The importance of demographic processes affecting inequality is underlined, and the impact of severe mortality crises, like the Black Death, is analyzed. Keywords Economic inequality; wealth concentration; middle ages; early modern period; Piedmont; Sabaudian States; Italy; plague; Black Death Acknowledgements The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement No. 283802, EINITE-Economic Inequality across Italy and Europe, 1300-1800. 2 The history of long-term trends in economic inequality is still largely to be written. After decades of research, mostly generated by Simon Kuznets' seminal 1955 article which introduced the notion that inequality would follow an inverted-U path through the industrialization process (the so-called ‘Kuznets curve’)1, we now have a good knowledge of the developments in inequality through the last couple of centuries, although only for selected countries like Britain, France, Italy, Spain, and the U.S. -

Settore Studi, Statistica E Documentazione Camera Di Commercio Di Torino
Torino, 3 aprile 2008 Settore Studi, Statistica e Documentazione Camera di commercio di Torino Nati-mortalità delle imprese della provincia di Torino nel 2007 CAMERA DI COMMERCIO INDUSTRIA ARTIGIANATO E AGRICOLTURA DI TORINO Settore Studi, Statistica e Documentazione Torino, 3 aprile 2008 Alessandro Barberis Presidente Camera di commercio di Torino CAMERA DI COMMERCIO INDUSTRIA ARTIGIANATO E AGRICOLTURA DI TORINO Settore Studi, Statistica e Documentazione Torino, 3 aprile 2008 Tassi di crescita del tessuto imprenditoriale: anni 2000-2007 2,0% 1,8% 1,6% 1,4% Torino: 1,32% 1,2% 1,0% Nord Ovest: 0,99% 0,8% Italia: 0,75% Piemonte: 0,69% 0,6% 0,4% 0,2% 0,0% 2000 2001 2002 2003 2004 2005 2006* 2007* (*) al netto delle cancellazioni d’ufficio Fonte: elaborazione Camera di commercio di Torino su dati InfoCamere CAMERA DI COMMERCIO INDUSTRIA ARTIGIANATO E AGRICOLTURA DI TORINO Settore Studi, Statistica e Documentazione Torino, 3 aprile 2008 Imprese registrate per settori al 31.12.2007 Totale imprese : 234.409 Imprese Agricoltura 50.168 Servizi 51.916non classificate e pesca alla persona 6,6% 6,2% Servizi 5,0% Turismo alle imprese 4,8% 23,7% 173.862 175.728 Commercio 26,6% Industria 11,9% Costruzioni 15,3% Fonte: elaborazione della Camera di commercio di Torino su dati InfoCamere CAMERA DI COMMERCIO INDUSTRIA ARTIGIANATO E AGRICOLTURA DI TORINO Settore Studi, Statistica e Documentazione Torino, 3 aprile 2008 Le 9 aree sub-provinciali (percentuale delle imprese sul totale) 11,0% Ivrea Po 5,6% Susa Sangone Susa Chivasso Bardonecchia 3,2% Stura 7,6% -

Piedmont Region Case Study
ReSSI – Regional strategies for sustainable and inclusive territorial development – Regional interplay and EU dialogue Targeted Analysis Annex 5 – Piedmont Region Case Study Version 30/11/2017 This targeted analysis activity is conducted within the framework of the ESPON 2020 Cooperation Programme, partly financed by the European Regional Development Fund. The ESPON EGTC is the Single Beneficiary of the ESPON 2020 Cooperation Programme. The Single Operation within the programme is implemented by the ESPON EGTC and co-financed by the European Regional Development Fund, the EU Member States and the Partner States, Iceland, Liechtenstein, Norway and Switzerland. This delivery does not necessarily reflect the opinion of the members of the ESPON 2020 Monitoring Committee. Authors Giancarlo Cotella, Elena Pede and Marco Santangelo, Interuniversity Department of Regional and Urban Studies and Planning – Politecnico di Torino (Italy) Advisory Group ESPON EGTC: Michaela Gensheimer (Senior Project Expert, Cluster Coordinator for Project Development and Coordination), Piera Petruzzi (Senior Project Expert, Communication and Capitalisation), Johannes Kiersch (Financial Expert) Information on ESPON and its projects can be found on www.espon.eu. The web site provides the possibility to download and examine the most recent documents produced by finalised and ongoing ESPON projects. This delivery exists only in an electronic version. © ESPON, 2017 Printing, reproduction or quotation is authorised provided the source is acknowledged and a copy is forwarded to the ESPON EGTC in Luxembourg. Contact: [email protected] ISBN: 978-99959-55-16-8 a ReSSI Regional strategies for sustainable and inclusive territorial development – Regional interplay and EU dialogue Table of contents List of Figures ............................................................................................................................. ii List of Tables ............................................................................................................................. -

Press Release Corner Rosa and Loans for Female
PRESS RELEASE CORNER ROSA AND LOANS FOR FEMALE ENTREPRENEURS • Intesa Sanpaolo is carrying out a trial in Piedmont offering dedicated help- desks for self-employed women and young people • Unsecured loans of up to 40,000 Euros available Turin/Milan, 10 March 2008 – Intesa Sanpaolo intends to facilitate access to credit for female entrepreneurs and young people. This will start in Piedmont, where there is a particularly high level of interest due to the establishment of a special guarantee fund and the memorandum of understanding signed in December between the Region, Unioncamer e and the regional ABI Commission. The Bank is making available loans which are backed 80% by a guarantee issued by FinPiemonte against the regional fund. These are initiatives targeted at women with no age limits and young people between 18 and 35 who already run or who are setting up their own business. They can be used to set up systems, make new investments and refurbish premises. The amount varies between 5,000 and 40,000 Euros repayable in 60 months at most. No further personal guarantees or collateral are required. The loans may be requested at all the Piedmont branches of the Intesa Sanpaolo Group. In order to provide applicants with a particularly focused advisory service, the Bank has created the Corner Rosa . These are areas that are instantly recognisable inside the branch, supervised by women who have followed a training course designed to give advice to other women and young people getting to grips with running a business. There are 20 Corner Rosa , located in Turin, Chieri, Rivoli, Pinerolo, Ivrea, Borgomanero, Omegna, Verbania, Novara, Vercelli, Biella, Alessandria, Asti and Cuneo, which have been operating since 8 March. -

GIRONE a GIRONE B Volpiano Venaria Arancione Lascaris Chieri
XIII GRAN GALA' DELLA SCUOLA CALCIO CATEGORIA PRIMI CALCI 2013 (TRE TEMPI DA 12 MINUTI) CAMPO VENARIA REALE - VIA SAN MARCHESE 27, VENARIA GIRONE A GIRONE B Volpiano Venaria Arancione Lascaris Chieri Pinerolo Garino Ciriè Lucento GIRONE C GIRONE D Chisola Mirafiori Borgaro Venaria Verde Rivarolese Lenci Poirino Cenisia Sisport DATA ORA GIRONE A - INCONTRO RISULTATO NOTE 12/09/2020 15:00 Volpiano Lascaris 12/09/2020 15:00 Pinerolo Ciriè 13/09/2020 15:00 Lascaris Pinerolo 13/09/2020 15:00 Ciriè Volpiano 19/09/2020 15:00 Volpiano Pinerolo 19/09/2020 15:00 Lascaris Ciriè DATA ORA GIRONE B - INCONTRO RISULTATO NOTE 12/09/2020 16:00 Venaria Arancione Chieri 12/09/2020 16:00 Garino Lucento 13/09/2020 16:00 Chieri Garino 13/09/2020 16:00 Lucento Venaria Arancione 19/09/2020 16:00 Venaria Arancione Garino 19/09/2020 16:00 Chieri Lucento DATA ORA GIRONE C - INCONTRO RISULTATO NOTE 12/09/2020 17:00 Chisola Borgaro 12/09/2020 17:00 Rivarolese Cenisia 20/09/2020 15:00 Borgaro Rivarolese 13/09/2020 17:00 Cenisia Chisola 19/09/2020 17:00 Chisola Rivarolese 19/09/2020 17:00 Borgaro Cenisia DATA ORA GIRONE D - INCONTRO RISULTATO NOTE 12/09/2020 18:00 Mirafiori Venaria Verde 12/09/2020 18:00 Lenci Poirino Sisport 13/09/2020 18:00 Venaria Verde Lenci Poirino 20/09/2020 16:00 Sisport Mirafiori 19/09/2020 18:00 Mirafiori Lenci Poirino 19/09/2020 18:00 Venaria Verde Sisport GIRONE E GIRONE F Volpiano Lascaris Venaria Arancione Chieri Chisola Borgaro Mirafiori Venaria Verde GIRONE G GIRONE H Pinerolo Ciriè Garino Lucento Rivarolese Cenisia Lenci Poirino Sisport -

Uffici Locali Dell'agenzia Delle Entrate E Competenza
TORINO Le funzioni operative dell'Agenzia delle Entrate sono svolte dalle: Direzione Provinciale I di TORINO articolata in un Ufficio Controlli, un Ufficio Legale e negli uffici territoriali di MONCALIERI , PINEROLO , TORINO - Atti pubblici, successioni e rimborsi Iva , TORINO 1 , TORINO 3 Direzione Provinciale II di TORINO articolata in un Ufficio Controlli, un Ufficio Legale e negli uffici territoriali di CHIVASSO , CIRIE' , CUORGNE' , IVREA , RIVOLI , SUSA , TORINO - Atti pubblici, successioni e rimborsi Iva , TORINO 2 , TORINO 4 La visualizzazione della mappa dell'ufficio richiede il supporto del linguaggio Javascript. Direzione Provinciale I di TORINO Comune: TORINO Indirizzo: CORSO BOLZANO, 30 CAP: 10121 Telefono: 01119469111 Fax: 01119469272 E-mail: [email protected] PEC: [email protected] Codice Ufficio: T7D Competenza territoriale: Circoscrizioni di Torino: 1, 2, 3, 8, 9, 10. Comuni: Airasca, Andezeno, Angrogna, Arignano, Baldissero Torinese, Bibiana, Bobbio Pellice, Bricherasio, Buriasco, Cambiano, Campiglione Fenile, Cantalupa, Carignano, Carmagnola, Castagnole Piemonte, Cavour, Cercenasco, Chieri, Cumiana, Fenestrelle, Frossasco, Garzigliana, Inverso Pinasca, Isolabella, La Loggia, Lombriasco, Luserna San Giovanni, Lusernetta, Macello, Marentino, Massello, Mombello di Torino, Moncalieri, Montaldo Torinese, Moriondo Torinese, Nichelino, None, Osasco, Osasio, Pancalieri, Pavarolo, Pecetto Torinese, Perosa Argentina, Perrero, Pinasca, Pinerolo, Pino Torinese, Piobesi Torinese, Piscina, Poirino, Pomaretto, -

Collina Torinese
COLLINA TORINESE DISCIPLINARE DI PRODUZIONE DOC COLLINA PROPOSTA DI MODIFICA TORINESE Articolo 1 – (Riconoscimento denominazione) Art.1. Denominazione e vini La denominazione d’origine controllata Collina Torinese è La denominazione d’origine controllata Collina Torinese è riservata ai vini che rispondono alle condizioni ed ai requisiti riservata ai vini che rispondono alle condizioni ed ai requisiti stabiliti nel presente disciplinare di produzione per le stabiliti nel presente disciplinare di produzione per le seguenti tipologie: seguenti tipologie: Collina Torinese rosso; Collina Torinese rosso; Collina Torinese barbera; Collina Torinese barbera; Collina Torinese bonarda; Collina Torinese bonarda; Collina Torinese Malvasia; Collina Torinese Malvasia; Collina Torinese Pelaverga o Cari. Collina Torinese Pelaverga o Cari Collina Torinese rosso Novello. Articolo 2 – (Vitigni ammessi) I vini a denominazione d.o.c. Collina Torinese devono Art. 2. Base ampelografica essere ottenuti dalle uve prodotte dai vigneti aventi, 1. I vini a denominazione d.o.c. Collina Torinese devono nell’ambito aziendale, la seguente composizione essere ottenuti dalle uve prodotte dai vigneti aventi, ampelografica: nell’ambito aziendale, la seguente composizione ampelografica: Collina Torinese rosso: Barbera : minimo 60%; Collina Torinese rosso e Collina Torinese rosso Novello: Freisa: minimo 25%, Barbera : minimo 60%; possono concorrere alla produzione di detto vino altri Freisa: minimo 25%, vitigni a bacca rossa, non aromatici, raccomandati e/o possono concorrere -

CURRICULUM PROFESSIONALE Dr. Roberto RECUPERO (Dichiarazione Sostitutiva Di Certificazione - Art
CURRICULUM PROFESSIONALE Dr. Roberto RECUPERO (dichiarazione sostitutiva di certificazione - art. 46 e dell'atto di notorietà - art. 47 e art.38 del D.P.R. 445/2000) Il sottoscritto Roberto Recupero nato il 13 agosto 1957 a Torino (prov. di TO) residente a Pino Torinese (TO) in Via Tepice n. 24 consapevole che in caso di dichiarazioni mendaci o non più rispondenti a verità e di formazione o uso di atti falsi, si applicheranno le sanzioni penali richiamate dall’art. 76 del vigente Testo unico delle disposizioni legislative e regolamentari in materia di documentazione amministrativa e che, ai sensi dell’art. 75 del Testo Unico citato decadrà dai benefici eventualmente conseguenti al provvedimento emanato sulla base della dichiarazione non veritiera, sotto la sua personale responsabilità DICHIARA di essere in possesso della Laurea in Medicina e Chirurgia conseguita in data 29/10/1982 presso l’Università degli Studi di Torino, con votazione: 110/110 lode e dignità di stampa di essere iscritto all’albo dell’ordine dei medici della Provincia di Torino dal 25/01/1983 n° di iscrizione 12301 di essere in possesso dei seguenti diplomi di specializzazione: 1) Medicina Interna indirizzo Medicina d'Urgenza conseguito in data 12/07/1991 presso l’Università di Torino normativa antecedente il DLgs 257/1991 durata anni 5 2 ) Endocrinologia conseguito in data 3/07/1985 presso l’Università di Torino normativa antecedente il DLgs 257/1991 durata anni 3 - di aver conseguito le seguenti idoneità alle funzioni di Direttore di Struttura Complessa di Medicina e Chirurgia d'Accettazione e d'Urgenza ( in accordo con il d.l. -

Linking Multifunctionality and Sustainability for Valuing Peri-Urban Farming: a Case Study in the Turin Metropolitan Area (Italy)
sustainability Article Linking Multifunctionality and Sustainability for Valuing Peri-Urban Farming: A Case Study in the Turin Metropolitan Area (Italy) Paola Gullino 1 ID , Luca Battisti 1 and Federica Larcher 1,2,* ID 1 Department of Agricultural, Forest and Food Sciences, University of Turin, Largo Paolo Braccini 2, 10095 Grugliasco (Turin), Italy; [email protected] (P.G.); [email protected] (L.B.) 2 Research Centre for Rural Development of Hilly, University of Turin, Largo Paolo Braccini 2, 10095 Grugliasco (Turin), Italy * Correspondence: [email protected]; Tel.: +39-011-6708-793 Received: 16 April 2018; Accepted: 14 May 2018; Published: 18 May 2018 Abstract: Agriculture plays a key role in managing the peri-urban landscapes in Europe, influencing their social, aesthetic and environmental functions. Considering the increase in urban population and land consumption in the last decades, sustainability in peri-urban areas is a priority. Farming multifunctionality is the integration of different functions and activities that produce beneficial effects on local economy, environment and society. Three research questions were explored: How is multifunctionality applied in peri-urban agroecosystems? How do we ensure sustainability in peri-urban agroecosystem? How could a bottom-up approach promote sustainable actions, strategies and policies? The Chieri Municipality (Turin Metropolitan Area, Italy) was chosen as representative case study. A trans-scalar approach from the farm to the municipality levels was adopted. The analysis of statistical data and farmers’ interviews were performed. Multifunctionality for three main farm categories (crops and grasslands; vineyards and orchards; and horticulture) was explored using the following parameters: website presence, online selling, agritourism, didactic farms, nonagricultural activities, maintenance parks and gardens, renewable energy, and transformation.