Figure4 July 14
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Production of Muconic Acid in Plants T ⁎ Aymerick Eudesa,B, , Roland Berthomieua,C, Zhangying Haoa,B, Nanxia Zhaoa,D, ⁎ Veronica Teixeira Benitesa,E, Edward E.K
Metabolic Engineering 46 (2018) 13–19 Contents lists available at ScienceDirect Metabolic Engineering journal homepage: www.elsevier.com/locate/meteng Production of muconic acid in plants T ⁎ Aymerick Eudesa,b, , Roland Berthomieua,c, Zhangying Haoa,b, Nanxia Zhaoa,d, ⁎ Veronica Teixeira Benitesa,e, Edward E.K. Baidooa,e, Dominique Loquéa,b,f,g, a Joint BioEnergy Institute, EmeryStation East, 5885 Hollis St, 4th Floor, Emeryville, CA 94608, USA b Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA c Ecole Polytechnique, Université Paris-Saclay, Palaiseau 91120, France d Department of Bioengineering, Department of Chemical & Biomolecular Engineering, University of California, Berkeley, CA 94720, USA e Biological Systems and Engineering Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA f Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720, USA g Université Lyon 1, INSA de Lyon, CNRS, UMR5240, Microbiologie, Adaptation et Pathogénie, 10 rue Raphaël Dubois, F-69622, Villeurbanne, France ARTICLE INFO ABSTRACT Keywords: Muconic acid (MA) is a dicarboxylic acid used for the production of industrially relevant chemicals such as Muconic acid adipic acid, terephthalic acid, and caprolactam. Because the synthesis of these polymer precursors generates Salicylic acid toxic intermediates by utilizing petroleum-derived chemicals and corrosive catalysts, the development of al- Catechol ternative strategies for the bio-based production of MA has garnered significant interest. Plants produce organic Shikimate carbon skeletons by harvesting carbon dioxide and energy from the sun, and therefore represent advantageous Plastid hosts for engineered metabolic pathways towards the manufacturing of chemicals. -

Medications in Pregnant and Nursing Mothers
Medications in Pregnant and Nursing Mothers NADINE M. GIRGIS, OD, FAAO ASSISTANT PROFESSOR YIN C. TEA, OD, FAAO CHIEF, PEDIATRICS AND BINOCULAR VISION ASSISTANT PROFESSOR Gestation age vs fetal age Gestation age-sperm penetrates the egg and zygote is formed Zygote (fertilized egg) travels from fallopian tube to uterus During this time, egg divides into cells - called a morula Continued dividing and morula - called a blastocyst - embeds in the uterus anywhere from 6-12 days after conception This begins the embryonic stage and fetal age begins Fetal development-1st trimester Gestation age week 3-fetal age week 1: a lot of basic growth Brain, spinal cord, heart, GI tract begin development 1st trimester Gestation age-week 4 and 5: embryo ¼ inch long Arm and leg buds, ears, eyes forming Placenta forming and producing hormones Heart is beating at a steady rhythm Movement of rudimentary blood through blood vessels 1st trimester Gestation age week 6: embryo is ½ in length Lungs, jaw, nose, plate formation, hands and feet Hand and feet buds have webbed structures Brain forming into complex parts 1st trimester Gestation age week 7: weighs less than an aspirin All essential organs have begun to form Hair, nail follicles, eyelids and tongue starting to form Trunk begins to straighten out 1st trimester Gestation age week 8: 1 in long, size of a bean All parts of adult are now present in the embryo Bones beginning to form Muscles begin to contract Facial features, including eyelids more developed Gestation age weeks 9-13: 3 in and weighs -

Genetic Characterisaton of Rhodococcus Rhodochrous ATCC
The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town Genetic characterization of Rhodococcus rhodochrous ATCC BAA-870 with emphasis on nitrile hydrolysing enzymes n ow Joni Frederick A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy in the Departmentty of of MolecularCape and T Cell Biology, Universitysi of Cape Town er UnivSupervisor: Professor B. T. Sewell Co-supervisor: Professor D. Brady February 2013 Keywords Nitrile hydrolysis Biocatalysis Rhodococcus rhodochrous ATCC BAA-870 Genome sequencing Nitrilase Nitrile hydratase n ow ty of Cape T si er Univ ii Keywords Abstract Rhodococcus rhodochrous ATCC BAA-870 (BAA-870) had previously been isolated on selective media for enrichment of nitrile hydrolysing bacteria. The organism was found to have a wide substrate range, with activity against aliphatics, aromatics, and aryl aliphatics, and enantioselectivity towards beta substituted nitriles and beta amino nitriles, compounds that have potential applications in the pharmaceutical industry. This makes R. rhodochrous ATCC BAA-870 potentially a versatile biocatalyst for the synthesis of a broad range of compounds with amide and carboxylic acid groups that can be derived from structurally related nitrile precursors. The selectivity of biocatalysts allows for high product yields and better atom economyn than non- selective chemical methods of performing this reaction, suchow as acid or base hydrolysis. -

Crystal Structure of Chorismate Synthase: a Novel FMN-Binding Protein Fold and Functional Insights
doi:10.1016/j.jmb.2003.12.072 J. Mol. Biol. (2004) 336, 903–915 Crystal Structure of Chorismate Synthase: A Novel FMN-binding Protein Fold and Functional Insights Hyung Jun Ahn, Hye-Jin Yoon, Byung Il Lee and Se Won Suh* Department of Chemistry Chorismate synthase catalyzes the conversion of 5-enolpyruvylshikimate College of Natural Sciences 3-phosphate to chorismate in the shikimate pathway, which represents Seoul National University an attractive target for discovering antimicrobial agents and herbicides. Seoul 151-0742, South Korea Chorismate serves as a common precursor for the synthesis of aromatic amino acids and many aromatic compounds in microorganisms and plants. Chorismate synthase requires reduced FMN as a cofactor but the catalyzed reaction involves no net redox change. Here, we have deter- mined the crystal structure of chorismate synthase from Helicobacter pylori in both FMN-bound and FMN-free forms. It is a tetrameric enzyme, with each monomer possessing a novel “b-a-b sandwich fold”. Highly con- served regions, including several flexible loops, cluster together around the bound FMN to form the active site. The unique FMN-binding site is formed largely by a single subunit, with a small contribution from a neighboring subunit. The isoalloxazine ring of the bound FMN is signifi- cantly non-planar. Our structure illuminates the essential functional roles played by the cofactor. q 2004 Elsevier Ltd. All rights reserved. Keywords: aroC; chorismate synthase; FMN-binding protein; Helicobacter *Corresponding author pylori; shikimate -

Molecular Targets for Antifungals in Amino Acid and Protein Biosynthetic Pathways
Amino Acids https://doi.org/10.1007/s00726-021-03007-6 REVIEW ARTICLE Molecular targets for antifungals in amino acid and protein biosynthetic pathways Aleksandra Kuplińska1 · Kamila Rząd1 Received: 1 March 2021 / Accepted: 17 May 2021 © The Author(s) 2021 Abstract Fungi cause death of over 1.5 million people every year, while cutaneous mycoses are among the most common infections in the world. Mycoses vary greatly in severity, there are long-term skin (ringworm), nail or hair infections (tinea capitis), recurrent like vaginal candidiasis or severe, life-threatening systemic, multiorgan infections. In the last few years, increas- ing importance is attached to the health and economic problems caused by fungal pathogens. There is a growing need for improvement of the availability of antifungal drugs, decreasing their prices and reducing side efects. Searching for novel approaches in this respect, amino acid and protein biosynthesis pathways appear to be competitive. The route that leads from amino acid biosynthesis to protein folding and its activation is rich in enzymes that are descriptive of fungi. Blocking the action of those enzymes often leads to avirulence or growth inhibition. In this review, we want to trace the principal processes of fungi vitality. We present the data of genes encoding enzymes involved in amino acid and protein biosynthesis, potential molecular targets in antifungal chemotherapy, and describe the impact of inhibitors on fungal organisms. Keywords Antifungal targets · Amino acid biosynthesis · Protein biosynthesis · Candida · Aspergillus · Plant pathogens Introduction is approximately 30–55% and 50–100% for aspergillosis (Brown et al. 2012; Verweij et al. 2016; Haidar and Singh Invasive fungal mycoses may afect over 300 million peo- 2018). -

Analysis of Allosteric Communication Pathways in Class I Glutamine Amidotransferases
Analysis of Allosteric Communication Pathways in Class I Glutamine Amidotransferases Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der Fakultät für Biologie und vorklinische Medizin der Universität Regensburg vorgelegt von Florian Semmelmann aus Deggendorf Juli 2019 Das Promotionsgesuch wurde eingereicht am: 12.07.2019 Die Arbeit wurde angeleitet von: Prof. Dr. Reinhard Sterner Unterschrift: .............................................. Florian Semmelmann Die vorliegende Arbeit wurde im Zeitraum von Oktober 2015 bis Juli 2019 am Lehrstuhl Bio- chemie II des Institutes für Biophysik und Physikalische Biochemie der Fakultät für Biologie und Vorklinische Medizin der Universität Regensburg unter Leitung von Prof. Dr. Reinhard Sterner angefertigt. Diese Arbeit wurde vom 01.01.2016 bis zum 31.12.2018 durch ein Promotionsstipendium der Konrad-Adenauer-Stiftung gefördert. Abstract Class I glutamine amidotransferases (GATases) represent a ubiquitous family of enzymes that catalyze the incorporation of ammonia within various metabolic pathways. GATases are het- eromeric enzyme complexes consisting of at least one pair of glutaminase and synthase subunits in various oligomeric associations. The activities of the glutaminase and synthase subunits are allosterically coupled in most class I GATases in a way that the presence of substrate at the synthase subunit stimulates glutamine hydrolysis at the glutaminase active site. The structural basis of allosteric communication between both subunits is, however, not well understood. The first part of this thesis focuses on the molecular basis of allosteric coupling in 4-amino-4- deoxychorismate synthase (ADCS), an important but hitherto sparsely characterized member of class I GATases. Using ancestral sequence reconstruction, we generated a thermostable glu- taminase subunit, anc2PabA, solved its crystal structure by molecular replacement and used it to compute for the first time a reliable model of the E. -

Preferred Drug List
October 2021 Preferred Drug List The Preferred Drug List, administered by CVS Caremark® on behalf of Siemens, is a guide within select therapeutic categories for clients, plan members and health care providers. Generics should be considered the first line of prescribing. If there is no generic available, there may be more than one brand-name medicine to treat a condition. These preferred brand-name medicines are listed to help identify products that are clinically appropriate and cost-effective. Generics listed in therapeutic categories are for representational purposes only. This is not an all-inclusive list. This list represents brand products in CAPS, branded generics in upper- and lowercase Italics, and generic products in lowercase italics. PLAN MEMBER HEALTH CARE PROVIDER Your benefit plan provides you with a prescription benefit program Your patient is covered under a prescription benefit plan administered administered by CVS Caremark. Ask your doctor to consider by CVS Caremark. As a way to help manage health care costs, prescribing, when medically appropriate, a preferred medicine from authorize generic substitution whenever possible. If you believe a this list. Take this list along when you or a covered family member brand-name product is necessary, consider prescribing a brand name sees a doctor. on this list. Please note: Please note: • Your specific prescription benefit plan design may not cover • Generics should be considered the first line of prescribing. certain products or categories, regardless of their appearance in • This drug list represents a summary of prescription coverage. It is this document. Products recently approved by the U.S. Food and not all-inclusive and does not guarantee coverage. -

Specific Pattern of Gene Expression During Induction of Mouse Erythroleukemia Cells
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Specific pattern of gene expression during induction of mouse erythroleukemia cells Peter J. Fraser and Peter J. Curtis The Wistar Institute of Anatomy and Biology, Philadelphia, Pennsylvania 19104 USA We have studied the expression of several characterized genes during induction of mouse erythroleukemia (MEL) cells with dimethyl sulfoxide (DMSO) and have observed a specific pattern of changes in transcriptional activity and steady-state RNA levels associated with erythroid differentiation. During induction there is a gradual, steady decrease in total transcriptional activity and RNA content per cell, which by day 3 of DMSO treatment amounts to less than 50% of the level in the uninduced cell. During this time we observe increases in transcriptional activity for 5-aminolevulinic acid synthase, carbonic anhydrase form II, and band 3 coordinate with the large increase in [3-globin gene transcription. The results also demonstrate an early decrease in transcription for carbonic anhydrase form I, which precedes decreases in transcription for glyceraldehyde phosphate dehydrogenase and rRNA genes. Changes in steady-state RNA levels reflected changes in transcriptional activity during induction except for carbonic anhydrase II mRNA. These results represent the first report characterizing the regulated expression at transcriptional and posttranscriptional levels of several known genes that are characteristically expressed in the erythrocyte. The results demonstrate that coordinate gene expression in erythroid differentiation occurs primarily at the level of transcription. [Key Words: Coordinated gene regulation; transcriptional activity; erythroid differentiation] Received June 25, 1987; revised version accepted August 14, 1987. Adult erythroid differentiation requires activation and/ dinately such that they may assemble to form the char- or modulation of many genes whose expression is regu- acteristic erythrocyte cytoskeleton. -
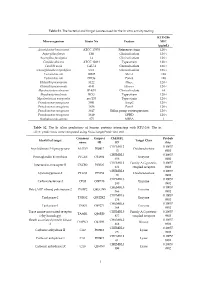
Table S1. the Bacterial and Fungal Isolates Used for the in Vitro Activity Testing
Table S1. The bacterial and fungal isolates used for the in vitro activity testing. KTU-286 Microorganism Strain No Feature MIC (µg/mL) Acinetobacter baummanii ATCC 17978 Reference strain 128 < Aspergillus flavus 12B Clinical isolate 128 < Aspergillus fumigatus 14 Clinical isolate 128 < Candida albicans ATCC 10231 Type strain 128 < Candida auris CAU-1 Clinical isolate 128 < Cuninghamella corymbifera CC1 Clinical isolate 128 < Escherichia coli 10025 Mcr-1 128 Escherichia coli DH5a Pan-S 128 Klebsiella pneumoniae 3122 blaKPC 128 < Klebsiella pneumoniae 4141 blaNDM-1 128 < Mycobacterium abscesus IP-K01 Clinical isolate 64 Mycobacterium bovis BCG Type strain 128 < Mycobacterium smegmatis mc2155 Type strain 128 < Pseudomonas aeruginosa 3691 AmpC 128 < Pseudomonas aeruginosa 3656 Pan-S 128 < Pseudomonas aeruginosa 3647 Efflux pump overexpression 128 < Pseudomonas aeruginosa 3619 OPRD 128 < Staphylococcus aureus 875 MRSA 1 Table S2. The In silico predictions of human proteins interacting with KTU-286. The in silico predictions were computed using SwissTargetPrediction tool. Common Uniprot ChEMBL Probab Identified target Target Class name ID ID ility CHEMBL2 0.10057 Arachidonate 5-lipoxygenase ALOX5 P09917 Oxidoreductase 15 8902 CHEMBL5 0.10057 Prostaglandin E synthase PTGES O14684 Enzyme 658 8902 CHEMBL2 Family A G protein- 0.10057 Interleukin-8 receptor B CXCR2 P25025 434 coupled receptor 8902 CHEMBL2 0.10057 Cyclooxygenase-2 PTGS2 P35354 Oxidoreductase 30 8902 CHEMBL3 0.10057 Carboxylesterase 2 CES2 O00748 Enzyme 180 8902 CHEMBL5 0.10057 Poly -

Supplementary Material Toxicological Impacts and Likely Protein Targets Of
Supplementary Material Toxicological impacts and likely protein targets of bisphenol A in Paramecium caudatum Marcus V. X. Senra† & Ana Lúcia Fonseca Instituto de Recursos Naturais, Universidade Federal de Itajubá, 37500-903, Itajubá, Minas Gerais – Brazil †To whom correspondence should be addressed – [email protected]; Orcid - 0000-0002-3866- 8837 Table S1. Annotation data on the P. caudatum 3D modelled proteins and their binding energies to BPA. BINDING ID DESCRIPTION CHROMOSOME NT_START NT_END ENERGIES (kcal/mol) PCAU.43c3d.1.P00010012 Tryptophan synthase beta subunit-like PLP-dependent enzyme scaffold_0001 23197 24853 -7.4 PCAU.43c3d.1.P00010015 Ribosomal protein L32e scaffold_0001 26373 26859 -6.2 PCAU.43c3d.1.P00010044 Catalase, mono-functional, haem-containing scaffold_0001 71821 73367 -6.5 PCAU.43c3d.1.P00010050 Dihydroorotate dehydrogenase, class 1/ 2 scaffold_0001 76614 79650 -6.6 PCAU.43c3d.1.P00010054 Serine/threonine/dual specificity protein kinase, catalytic domain scaffold_0001 83399 84653 -6.7 PCAU.43c3d.1.P00010070 Peptidyl-prolyl cis-trans isomerase, FKBP-type scaffold_0001 104909 105387 -5.9 PCAU.43c3d.1.P00010103 V-ATPase proteolipid subunit C-like domain scaffold_0001 168736 169346 -5.6 PCAU.43c3d.1.P00010112 DNA-directed RNA polymerase, RBP11-like dimerisation domain scaffold_0001 180310 181372 -6.0 PCAU.43c3d.1.P00010165 Vacuolar (H+)-ATPase G subunit scaffold_0001 252653 253112 -5.6 PCAU.43c3d.1.P00010176 Coproporphyrinogen III oxidase, aerobic scaffold_0001 262051 263168 -6.7 PCAU.43c3d.1.P00010205 Metalloenzyme,