Msx Homeobox Gene Family and Craniofacial Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Te2, Part Iii
TERMINOLOGIA EMBRYOLOGICA Second Edition International Embryological Terminology FIPAT The Federative International Programme for Anatomical Terminology A programme of the International Federation of Associations of Anatomists (IFAA) TE2, PART III Contents Caput V: Organogenesis Chapter 5: Organogenesis (continued) Systema respiratorium Respiratory system Systema urinarium Urinary system Systemata genitalia Genital systems Coeloma Coelom Glandulae endocrinae Endocrine glands Systema cardiovasculare Cardiovascular system Systema lymphoideum Lymphoid system Bibliographic Reference Citation: FIPAT. Terminologia Embryologica. 2nd ed. FIPAT.library.dal.ca. Federative International Programme for Anatomical Terminology, February 2017 Published pending approval by the General Assembly at the next Congress of IFAA (2019) Creative Commons License: The publication of Terminologia Embryologica is under a Creative Commons Attribution-NoDerivatives 4.0 International (CC BY-ND 4.0) license The individual terms in this terminology are within the public domain. Statements about terms being part of this international standard terminology should use the above bibliographic reference to cite this terminology. The unaltered PDF files of this terminology may be freely copied and distributed by users. IFAA member societies are authorized to publish translations of this terminology. Authors of other works that might be considered derivative should write to the Chair of FIPAT for permission to publish a derivative work. Caput V: ORGANOGENESIS Chapter 5: ORGANOGENESIS -

Interaction Network of Immune‑Associated Genes Affecting the Prognosis of Patients with Glioblastoma
EXPERIMENTAL AND THERAPEUTIC MEDICINE 21: 61, 2021 Interaction network of immune‑associated genes affecting the prognosis of patients with glioblastoma XIAOHONG HOU1*, JIALIN CHEN2*, QIANG ZHANG1, YINCHUN FAN1, CHENGMING XIANG1, GUIYIN ZHOU1, FANG CAO1 and SHENGTAO YAO1 1Department of Cerebrovascular Disease, Affiliated Hospital of Zunyi Medical University;2 Department of Neonatology, The First People' s Hospital of Zunyi Affiliated to Zunyi Medical University, Zunyi, Guizhou 563000, P.R. China Received October 15, 2019; Accepted October 6, 2020 DOI: 10.3892/etm.2020.9493 Abstract. Glioblastoma multiforme (GBM) is a common and immune genes of interest. The interaction network of malignant tumor type of the nervous system. The purpose immune‑regulatory genes constructed in the present study of the present study was to establish a regulatory network of enhances the current understanding of mechanisms associated immune‑associated genes affecting the prognosis of patients with poor prognosis of patients with GBM. The risk score with GBM. The GSE4290, GSE50161 and GSE2223 datasets model established in the present study may be used to evaluate from the Gene Expression Omnibus database were screened the prognosis of patients with GBM. to identify common differentially expressed genes (co‑DEGs). A functional enrichment analysis indicated that the co‑DEGs Introduction were mainly enriched in cell communication, regulation of enzyme activity, immune response, nervous system, cytokine Glioblastoma multiforme (GBM) is one of the most malig‑ signaling in immune system and the AKT signaling pathway. nant tumor types of the central nervous system, with short The co‑DEGs accumulated in immune response were then median survival and poor prognosis. -

Ncomms2254.Pdf
ARTICLE Received 3 May 2012 | Accepted 5 Nov 2012 | Published 4 Dec 2012 DOI: 10.1038/ncomms2254 The RB family is required for the self-renewal and survival of human embryonic stem cells Jamie F. Conklin1,2,3, Julie Baker2,3 & Julien Sage1,2,3 The mechanisms ensuring the long-term self-renewal of human embryonic stem cells are still only partly understood, limiting their use in cellular therapies. Here we found that increased activity of the RB cell cycle inhibitor in human embryonic stem cells induces cell cycle arrest, differentiation and cell death. Conversely, inactivation of the entire RB family (RB, p107 and p130) in human embryonic stem cells triggers G2/M arrest and cell death through functional activation of the p53 pathway and the cell cycle inhibitor p21. Differences in E2F target gene activation upon loss of RB family function between human embryonic stem cells, mouse embryonic stem cells and human fibroblasts underscore key differences in the cell cycle regulatory networks of human embryonic stem cells. Finally, loss of RB family function pro- motes genomic instability in both human and mouse embryonic stem cells, uncoupling cell cycle defects from chromosomal instability. These experiments indicate that a homeostatic level of RB activity is essential for the self-renewal and the survival of human embryonic stem cells. 1 Department of Pediatrics, Stanford Medical School, Stanford, California 94305, USA. 2 Department of Genetics, Stanford Medical School, Stanford, California 94305, USA. 3 Institute for Stem Cell Biology and Regenerative Medicine, Stanford Medical School, Stanford, California 94305, USA. Correspondence and requests for materials should be addressed to J.S. -

Selective Estrogen Receptor Modulators: Discrimination of Agonistic Versus Antagonistic Activities by Gene Expression Profiling in Breast Cancer Cells
[CANCER RESEARCH 64, 1522–1533, February 15, 2004] Selective Estrogen Receptor Modulators: Discrimination of Agonistic versus Antagonistic Activities by Gene Expression Profiling in Breast Cancer Cells Jonna Frasor,1 Fabio Stossi,1 Jeanne M. Danes,1 Barry Komm,2 C. Richard Lyttle,2 and Benita S. Katzenellenbogen1 1Department of Molecular and Integrative Physiology, University of Illinois and College of Medicine, Urbana, Illinois, and 2Women’s Health Research Institute, Wyeth Research, Collegeville, Pennsylvania ABSTRACT tures in these women; however, some detrimental side effects such as an increased risk of endometrial cancer, stroke, and pulmonary embolism Selective estrogen receptor modulators (SERMs) such as tamoxifen are were also associated with tamoxifen treatment (7). Ral was examined in effective in the treatment of many estrogen receptor-positive breast cancers the Multiple Outcomes of Raloxifene Evaluation trial and found to be and have also proven to be effective in the prevention of breast cancer in women at high risk for the disease. The comparative abilities of tamoxifen effective in reducing the incidence of osteoporosis in postmenopausal versus raloxifene in breast cancer prevention are currently being compared in women, as well as the incidence of breast cancer but, unlike tamoxifen, the Study of Tamoxifen and Raloxifene trial. To better understand the actions without the increased risk of endometrial cancer (8, 9). On the basis of the of these compounds in breast cancer, we have examined their effects on the positive outcome of these trials, the Study of Tamoxifen and Raloxifene expression of ϳ12,000 genes, using Affymetrix GeneChip microarrays, with trial was begun in 1999 to directly compare the effects of these two quantitative PCR verification in many cases, categorizing their actions as SERMs, tamoxifen and Ral, in prevention of breast cancer (10, 11). -

Visualizing Morphogenesis with the Processing Programming Language 15
Visualizing Morphogenesis with the Processing Programming Language 15 Visualizing Morphogenesis with the Processing Programming Language Avik Patel, Amar Bains, Richard Millet, and Tamira Elul changing tissue shapes. A powerful technique for elucidating the We used Processing, a visual artists’ programming dynamic cellular mechanisms of morphogenesis is time-lapse video- language developed at MIT Media Lab, to simulate microscopic imaging of cells in embryonic tissues undergoing cellular mechanisms of morphogenesis – the generation morphogenesis (Elul and Keller 2000; Elul et al., 1997; Harris et al., of form and shape in embryonic tissues. Based on 1987). The mechanisms underlying morphogenetic processes can be clarified by observation of cell dynamics from these time-lapse video observations of in vivo time-lapse image sequences, we sequences. Morphometric measurement further defines the created animations of neural cell motility responsible for quantitative and statistical relationships between the cell dynamics that elongating the spinal cord, and of optic axon branching underlie morphogenesis (Kim and Davidson 2011; Marshak et al., dynamics that establish primary visual connectivity. 2007; Elul et al., 1997; Witte et al., 1996). Mathematical These visual models underscore the significance of the decomposition of cell dynamics additionally facilitates the computational decomposition of cellular dynamics computational modeling of cellular mechanisms that drive underlying morphogenesis. morphogenesis (Satulovsky et al., 2008). Methods In this paper, we use the Processing programming language to visualize dynamic cell behaviors driving morphogenesis in the Introduction developing nervous system. Based on in vivo time-lapse image sequences, we created models of cell dynamics underlying two Processing is a Java based software language and environment morphogenetic processes in the developing nervous system. -

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B
Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B. M ü ller, G ü nter P. Wagner, and Werner Callebaut, editors The Evolution of Cognition , edited by Cecilia Heyes and Ludwig Huber, 2000 Origination of Organismal Form: Beyond the Gene in Development and Evolutionary Biology , edited by Gerd B. M ü ller and Stuart A. Newman, 2003 Environment, Development, and Evolution: Toward a Synthesis , edited by Brian K. Hall, Roy D. Pearson, and Gerd B. M ü ller, 2004 Evolution of Communication Systems: A Comparative Approach , edited by D. Kimbrough Oller and Ulrike Griebel, 2004 Modularity: Understanding the Development and Evolution of Natural Complex Systems , edited by Werner Callebaut and Diego Rasskin-Gutman, 2005 Compositional Evolution: The Impact of Sex, Symbiosis, and Modularity on the Gradualist Framework of Evolution , by Richard A. Watson, 2006 Biological Emergences: Evolution by Natural Experiment , by Robert G. B. Reid, 2007 Modeling Biology: Structure, Behaviors, Evolution , edited by Manfred D. Laubichler and Gerd B. M ü ller, 2007 Evolution of Communicative Flexibility: Complexity, Creativity, and Adaptability in Human and Animal Communication , edited by Kimbrough D. Oller and Ulrike Griebel, 2008 Functions in Biological and Artifi cial Worlds: Comparative Philosophical Perspectives , edited by Ulrich Krohs and Peter Kroes, 2009 Cognitive Biology: Evolutionary and Developmental Perspectives on Mind, Brain, and Behavior , edited by Luca Tommasi, Mary A. Peterson, and Lynn Nadel, 2009 Innovation in Cultural Systems: Contributions from Evolutionary Anthropology , edited by Michael J. O ’ Brien and Stephen J. Shennan, 2010 The Major Transitions in Evolution Revisited , edited by Brett Calcott and Kim Sterelny, 2011 Transformations of Lamarckism: From Subtle Fluids to Molecular Biology , edited by Snait B. -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
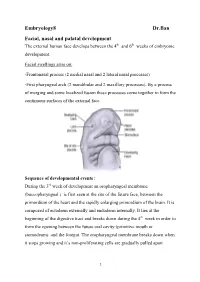
Embryology8 Dr.Ban Facial, Nasal and Palatal Development the External Human Face Develops Between the 4Th and 6Th Weeks of Embryonic Development
Embryology8 Dr.Ban Facial, nasal and palatal development The external human face develops between the 4th and 6th weeks of embryonic development. Facial swellings arise on: -Frontonasal process (2 medial nasal and 2 lateral nasal processes) -First pharyngeal arch (2 mandibular and 2 maxillary processes). By a process of merging and some localized fusion these processes come together to form the continuous surfaces of the external face. Sequence of developmental events : During the 3rd week of development an oropharyngeal membrane (buccopharyngeal ) is first seen at the site of the future face, between the primordium of the heart and the rapidly enlarging primordium of the brain. It is composed of ectoderm externally and endoderm internally. It lies at the beginning of the digestive tract and breaks down during the 4th week in order to form the opening between the future oral cavity (primitive mouth or stomodeum) and the foregut. The oropharyngeal membrane breaks down when it stops growing and it’s non-proliferating cells are gradually pulled apart 1 because they cannot fill the expanding area.The tissues around it expand very rapidly. The face develops from five primordia that appear in the fourth week: the frontonasal prominence, the two maxillary swellings, and the two mandibular swellings. The external face forms from two sources that surround the oropharyngeal membrane 1-Tissues of the frontonasal process that cover the forebrain, predominantly of neural crest origin. 2-The tissues of the first (or mandibular) pharyngeal arch, of mixed mesoderm and neural crest origin Face initially formed by 5 mesenchymal swellings ( prominences): Two mandibular prominences Two maxillary prominences Frontonasal prominence (midline structure is a single structure that is ventral to the forebrain. -

Expression of the Human PAC1 Receptor Leads to Dose- Dependent Hydrocephalus-Related Abnormalities in Mice
Expression of the human PAC1 receptor leads to dose- dependent hydrocephalus-related abnormalities in mice Bing Lang, … , Anthony J. Harmar, Sanbing Shen J Clin Invest. 2006;116(7):1924-1934. https://doi.org/10.1172/JCI27597. Research Article Neuroscience Hydrocephalus is a common and potentially devastating birth defect affecting the CNS, and its relationship with G protein–coupled receptors (GPCRs) is unknown. We have expressed 2, 4, or 6 copies of a GPCR — the human PAC1 receptor with a 130-kb transgene in the mouse nervous system in a pattern closely resembling that of the endogenous gene. Consistent with PAC1 actions, PKA and PKC activity were elevated in the brains of Tg mice. Remarkably, Tg mice developed dose-dependent hydrocephalus-like characteristics, including enlarged third and lateral ventricles and reduced cerebral cortex, corpus callosum, and subcommissural organ (SCO). Neuronal proliferation and apoptosis were implicated in hydrocephalus, and we observed significantly reduced neuronal proliferation and massively increased neuronal apoptosis in the developing cortex and SCO of Tg embryos, while neurite outgrowth and neuronal migration in vitro remain uncompromised. Ventricular ependymal cilia are crucial for directing cerebrospinal fluid flow, and ependyma of Tg mice exhibited disrupted cilia with increased phospho-CREB immunoreactivity. These data demonstrate that altered neuronal proliferation/apoptosis and disrupted ependymal cilia are the main factors contributing to hydrocephalus in PAC1-overexpressing mice. This is the first report to our knowledge demonstrating that misregulation of GPCRs can be involved in hydrocephalus-related neurodevelopmental disorders. Find the latest version: https://jci.me/27597/pdf Research article Related Commentary, page 1828 Expression of the human PAC1 receptor leads to dose-dependent hydrocephalus- related abnormalities in mice Bing Lang,1 Bing Song,1 Wendy Davidson,1 Alastair MacKenzie,1 Norman Smith,2 Colin D. -

Vocabulario De Morfoloxía, Anatomía E Citoloxía Veterinaria
Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) Servizo de Normalización Lingüística Universidade de Santiago de Compostela COLECCIÓN VOCABULARIOS TEMÁTICOS N.º 4 SERVIZO DE NORMALIZACIÓN LINGÜÍSTICA Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) 2008 UNIVERSIDADE DE SANTIAGO DE COMPOSTELA VOCABULARIO de morfoloxía, anatomía e citoloxía veterinaria : (galego-español- inglés) / coordinador Xusto A. Rodríguez Río, Servizo de Normalización Lingüística ; autores Matilde Lombardero Fernández ... [et al.]. – Santiago de Compostela : Universidade de Santiago de Compostela, Servizo de Publicacións e Intercambio Científico, 2008. – 369 p. ; 21 cm. – (Vocabularios temáticos ; 4). - D.L. C 2458-2008. – ISBN 978-84-9887-018-3 1.Medicina �������������������������������������������������������������������������veterinaria-Diccionarios�������������������������������������������������. 2.Galego (Lingua)-Glosarios, vocabularios, etc. políglotas. I.Lombardero Fernández, Matilde. II.Rodríguez Rio, Xusto A. coord. III. Universidade de Santiago de Compostela. Servizo de Normalización Lingüística, coord. IV.Universidade de Santiago de Compostela. Servizo de Publicacións e Intercambio Científico, ed. V.Serie. 591.4(038)=699=60=20 Coordinador Xusto A. Rodríguez Río (Área de Terminoloxía. Servizo de Normalización Lingüística. Universidade de Santiago de Compostela) Autoras/res Matilde Lombardero Fernández (doutora en Veterinaria e profesora do Departamento de Anatomía e Produción Animal.