Exploring the Biochemical and Phylogenetic Fingerprint of Australian Native Plants for Sustainable Use of Saline Lands
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materialsupplementary Material
10.1071/BT13149_AC © CSIRO 2013 Australian Journal of Botany 2013, 61(6), 436–445 SUPPLEMENTARY MATERIAL Comparative dating of Acacia: combining fossils and multiple phylogenies to infer ages of clades with poor fossil records Joseph T. MillerA,E, Daniel J. MurphyB, Simon Y. W. HoC, David J. CantrillB and David SeiglerD ACentre for Australian National Biodiversity Research, CSIRO Plant Industry, GPO Box 1600 Canberra, ACT 2601, Australia. BRoyal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Vic. 3141, Australia. CSchool of Biological Sciences, Edgeworth David Building, University of Sydney, Sydney, NSW 2006, Australia. DDepartment of Plant Biology, University of Illinois, Urbana, IL 61801, USA. ECorresponding author. Email: [email protected] Table S1 Materials used in the study Taxon Dataset Genbank Acacia abbreviata Maslin 2 3 JF420287 JF420065 JF420395 KC421289 KC796176 JF420499 Acacia adoxa Pedley 2 3 JF420044 AF523076 AF195716 AF195684; AF195703 Acacia ampliceps Maslin 1 KC421930 EU439994 EU811845 Acacia anceps DC. 2 3 JF420244 JF420350 JF419919 JF420130 JF420456 Acacia aneura F.Muell. ex Benth 2 3 JF420259 JF420036 JF420366 JF419935 JF420146 KF048140 Acacia aneura F.Muell. ex Benth. 1 2 3 JF420293 JF420402 KC421323 JQ248740 JF420505 Acacia baeuerlenii Maiden & R.T.Baker 2 3 JF420229 JQ248866 JF420336 JF419909 JF420115 JF420448 Acacia beckleri Tindale 2 3 JF420260 JF420037 JF420367 JF419936 JF420147 JF420473 Acacia cochlearis (Labill.) H.L.Wendl. 2 3 KC283897 KC200719 JQ943314 AF523156 KC284140 KC957934 Acacia cognata Domin 2 3 JF420246 JF420022 JF420352 JF419921 JF420132 JF420458 Acacia cultriformis A.Cunn. ex G.Don 2 3 JF420278 JF420056 JF420387 KC421263 KC796172 JF420494 Acacia cupularis Domin 2 3 JF420247 JF420023 JF420353 JF419922 JF420133 JF420459 Acacia dealbata Link 2 3 JF420269 JF420378 KC421251 KC955787 JF420485 Acacia dealbata Link 2 3 KC283375 KC200761 JQ942686 KC421315 KC284195 Acacia deanei (R.T.Baker) M.B.Welch, Coombs 2 3 JF420294 JF420403 KC421329 KC955795 & McGlynn JF420506 Acacia dempsteri F.Muell. -

Human-Mediated Introductions of Australian Acacias
Diversity and Distributions, (Diversity Distrib.) (2011) 17, 771–787 S EDITORIAL Human-mediated introductions of PECIAL ISSUE Australian acacias – a global experiment in biogeography 1 2 1 3,4 David M. Richardson *, Jane Carruthers , Cang Hui , Fiona A. C. Impson , :H Joseph T. Miller5, Mark P. Robertson1,6, Mathieu Rouget7, Johannes J. Le Roux1 and John R. U. Wilson1,8 UMAN 1 Centre for Invasion Biology, Department of ABSTRACT - Botany and Zoology, Stellenbosch University, MEDIATED INTRODUCTIONS OF Aim Australian acacias (1012 recognized species native to Australia, which were Matieland 7602, South Africa, 2Department of History, University of South Africa, PO Box previously grouped in Acacia subgenus Phyllodineae) have been moved extensively 392, Unisa 0003, South Africa, 3Department around the world by humans over the past 250 years. This has created the of Zoology, University of Cape Town, opportunity to explore how evolutionary, ecological, historical and sociological Rondebosch 7701, South Africa, 4Plant factors interact to affect the distribution, usage, invasiveness and perceptions of a Protection Research Institute, Private Bag globally important group of plants. This editorial provides the background for the X5017, Stellenbosch 7599, South Africa, 20 papers in this special issue of Diversity and Distributions that focusses on the 5Centre for Australian National Biodiversity global cross-disciplinary experiment of introduced Australian acacias. A Journal of Conservation Biogeography Research, CSIRO Plant Industry, GPO Box Location Australia and global. 1600, Canberra, ACT, Australia, 6Department of Zoology and Entomology, University of Methods The papers of the special issue are discussed in the context of a unified Pretoria, Pretoria 0002, South Africa, framework for biological invasions. -

National Parks and Wildlife Act 1972.PDF
Version: 1.7.2015 South Australia National Parks and Wildlife Act 1972 An Act to provide for the establishment and management of reserves for public benefit and enjoyment; to provide for the conservation of wildlife in a natural environment; and for other purposes. Contents Part 1—Preliminary 1 Short title 5 Interpretation Part 2—Administration Division 1—General administrative powers 6 Constitution of Minister as a corporation sole 9 Power of acquisition 10 Research and investigations 11 Wildlife Conservation Fund 12 Delegation 13 Information to be included in annual report 14 Minister not to administer this Act Division 2—The Parks and Wilderness Council 15 Establishment and membership of Council 16 Terms and conditions of membership 17 Remuneration 18 Vacancies or defects in appointment of members 19 Direction and control of Minister 19A Proceedings of Council 19B Conflict of interest under Public Sector (Honesty and Accountability) Act 19C Functions of Council 19D Annual report Division 3—Appointment and powers of wardens 20 Appointment of wardens 21 Assistance to warden 22 Powers of wardens 23 Forfeiture 24 Hindering of wardens etc 24A Offences by wardens etc 25 Power of arrest 26 False representation [3.7.2015] This version is not published under the Legislation Revision and Publication Act 2002 1 National Parks and Wildlife Act 1972—1.7.2015 Contents Part 3—Reserves and sanctuaries Division 1—National parks 27 Constitution of national parks by statute 28 Constitution of national parks by proclamation 28A Certain co-managed national -

Acacia Eriopoda X Monticola
WATTLE Acacias of Australia Acacia eriopoda Maiden & Blakely x Acacia monticola J.M.Black Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com B.R. Maslin Family Fabaceae Distribution This entity has a scattered distribution, mostly in the Kimberley region (N and S of Broome and E and S of Derby), but also recorded from “near Well 22” which is assumed to be on the Canning Stock Route on the N side of Lake Disappointment in the Little Sandy Desert. Specimens W.A.: 48 mi [77.2 km] from Broome towards Port Hedland, C.H. Gittins 1451 (PERTH); near Well 22, D. Hewitt 26 (PERTH); 57 mi (91 km) from Derby [on Great Northern Highway] towards Fitzroy Crossing, B.R. Maslin 2669 (PERTH); 46 km N of Broome on track to Coulomb Point, B.R. Maslin 4900 (PERTH); Dampier Downs Station (c. 150 km due SW of Derby), 49 km NW of homestead on station track to Broome (NW of Edgar Ranges), B.R. Maslin 7322 (DNA, PERTH). Notes The reticulately nerved phyllodes and Minni Ritchi or pseudo Minni Ritchi bark which is noted by some collectors suggests A. monticola is one of the parents of this putative hybrid. However, the second parent is less certain. At four locations the putative hybrid is recorded as occurring (usually at a low frequency) either within a population of A. eriopoda and A. monticola (C.H. Gittins 1451 and B.R. Maslin 2669 & 4900) and or within a population of A. monticola with A. eriopoda nearby (i.e. -
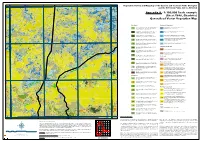
Appendix 9 - 1:100,000 Scale Example (Sheet 5648, Charlotte) Generalised Vector Vegetation Map
133°30'E 133°40'E 133°50'E 134°E 24°30'S Vegetation Survey and Mapping of the Eastern and Southern Finke Bioregion 24°30'S and the NT Stony Plains Inliers, NT & SA Appendix 9 - 1:100,000 Scale example (Sheet 5648, Charlotte) Generalised Vector Vegetation Map Woodland Chenopod Shrubland Acacia aneura ( Mulga) Low Open Woodland TO Tall Open Shrubland of Atriplex nummularia (Old man saltbush) Low Sparse Chenopod 1 Acacia estrophiolata (Ironwood) on clay loam plains and red earth 4 shrubland over Low Sparse Tussock grasses. soils+/- Atriplex vesicaria and Eragrostis eriopoda . Acacia georginae / Acacia cambagei ( Gidgee) Low Woodland to Tall Atriplex vesicaria (Pop saltbush) Low Open Chenopod Shrubland.+/- 2 Shrubland.+/- Eucalyptus coolabah subsp. Arida , Codonocarpus 5 Maireana astrotricha over tussock grasses. cotinifolius , Eulalia aurea, Eriachne ovata and Atriplex vesicaria . Eucalyptus coolabah subsp. arida (Coolabah) Woodland. +/- Maireana aphylla (Cottonbush) Low Sparse Chenopod Shrubland. +/- 12 Muehlenbeckia florulenta , Acacia aneura , Senna artemisioides ssp. 8 Fimbristylis dichotoma , Dactyloctenium radulans and Eragrostis dielsii. Filifolia , Marsilea sp ., Cynodon dactylon , and Cenchrus ciliaris . Maireana astrotricha (Low bluebush) Low Sparse Chenopod Shrubland Eucalyptus camaldulensis var. obtusa (River red gum) Woodland.+/- TO Sparse shrubland of Senna artemisioides n. coriacea and 13 Eucalyptus coolabah subsp. arida , Cynodon dactylon , Eulalia aurea and 9 Eremophila duttonii (Harlequin fuchsia bush). Cyperus gymnocaulos . 24°40'S Hakea leucoptera subsp. leucoptera (Needlewood) Open Woodland. +- Sclerolaena (mixed) Low Sparse Chenopod Shrubland.+/- Enneapogon 24°40'S 14 Eremophila sturtii , Senna artemisioides ssp. filifolia , Hakea leucoptera 15 avenaceus Aristida contorta , Sporobolus actinocladus . subsp. leucoptera and Triodia basedowii . Acacia calcicola (Northern Myall) Sparse Woodland +/- Eremophila Samphire Shrubland 23 duttonii , Acacia calcicola , Atriplex vesicaria , Maireana georgei and mixed short grasses. -

A Taxonomic Revision of Acacia Vernicifluaand A. Leprosa
A taxonomic revision of Acacia verniciflua and A. leprosa (Leguminosae: Mimosoideae) in Australia Bruce R. Maslin 1 and Daniel J. Murphy 2 1 Western Australian Herbarium, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, Western Australia 6983, Australia; e-mail: [email protected] 2 National Herbarium of Victoria, Royal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Victoria 3141, Australia; e-mail: [email protected] Introduction Abstract Acacia verniciflua (Varnish Wattle) and A. leprosa (Cinnamon Wattle) A revision of a taxonomically complex occur in temperate areas of eastern and southern Australia and as group of species allied to, and including, Acacia leprosa Sieber ex DC. hitherto defined were regarded as highly polymorphic species in need of and A. verniciflua A.Cunn. is presented. critical revision (Maslin 2001). The conventional separation between the These species predominate in species was the number of longitudinal nerves on their phyllodes, one in temperate regions of eastern Australia. A. leprosa and two in A. verniciflua, but as correctly noted by Court (1972, Two new species are described, p. 219), the importance of this character has been over-emphasised. A. rostriformis Maslin & D.J.Murphy and A. stictophylla Court ex Maslin During the nineteenth century a number of taxa were described that & D.J.Murphy and one, A. exudans were referable to the A. verniciflua– A. leprosa group, however, none Lindl., is resurrected. Acacia leprosa is of these names was ever taken up, presumably because of difficulties treated as a highly polymorphic species in defining the taxa and uncertainties concerning the application of comprising five varieties, four of which the names (some of which were based on plants cultivated in Europe are described as new, namely, A. -

Southern Gulf, Queensland
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

'Soils' and 'Vegetation'?
Is there a close association between ‘soils’ and ‘vegetation’? A case study from central western New South Wales M.O. Rankin1, 3, W.S Semple2, B.W. Murphy1 and T.B. Koen1 1 Department of Natural Resources, PO Box 445, Cowra, NSW 2794, AUSTRALIA 2 Department of Natural Resources, PO Box 53, Orange, NSW 2800, AUSTRALIA 3 Corresponding author, email: [email protected] Abstract: The assumption that ‘soils’ and ‘vegetation’ are closely associated was tested by describing soils and vegetation along a Travelling Stock Reserve west of Grenfell, New South Wales (lat 33° 55’S, long 147° 45’E). The transect was selected on the basis of (a) minimising the effects of non-soil factors (human interference, climate and relief) on vegetation and (b) the presence of various soil and vegetation types as indicated by previous mapping. ‘Soils’ were considered at three levels: soil landscapes (a broad mapping unit widely used in central western NSW), soil types (according to a range of classifications) and soil properties (depth, pH, etc.). ‘Vegetation’ was considered in three ways: vegetation type (in various classifications), density/floristic indices (density of woody species, abundance of native species, etc.) and presence/absence of individual species. Sites along the transect were grouped according to soil landscapes or soil types and compared to vegetation types or indices recorded at the sites. Various measures indicated low associations between vegetation types and soil landscapes or soil types. Except for infrequent occurrences of a soil type or landscape, any one soil type or landscape was commonly associated with a number of vegetation types and any one vegetation type was associated with a number of soil landscapes or soil types. -

MVG 16 Acacia Shrublands DRAFT
MVG 16 - ACACIA SHRUBLANDS Acacia hillii, Tanami Desert, NT (Photo: D. Keith) Overview The overstorey of MVG 16 is dominated by multi-stemmed acacia shrubs. The most widespread species is Acacia aneura (mulga). Mulga vegetation takes on a variety of structural expressions and is consequently classified partly within MVG 16 where the overstorey is dominated by multi-stemmed shrubs, partly within MVG 6 in accordance with the Kyoto Protocol definition of forest cover in Australia (trees > 2 m tall and crown cover > 20%, foliage projective cover > 10%); and partly within MVG 13 where the woody dominants are predominantly single-stemmed, but with crown cover less than 20%. Occurs where annual rainfall is below 250mm in southern Australia and below 350mm in northern Australia (Hodgkinson 2002; Foulkes et al. 2014). Species composition varies along rainfall gradients, with substrate and rainfall seasonality (Beadle 1981; Johnson and Burrows 1994). Transitions into MVG 13 Acacia woodlands with higher rainfall and varying soil types. Is most commonly found on red earth soils (Hodgkinson 2002). Facts and figures Major Vegetation Group MVG 16 - Acacia Shrublands Major Vegetation Subgroups 20. Stony mulga woodlands and shrublands NSW, (number of NVIS descriptions) NT, QLD, SA, WA 23. Sandplain Acacia woodlands and shrublands NSW, NT, QLD, SA, WA Typical NVIS structural formations Shrubland (tall, mid,) Open shrubland (tall, mid,) Sparse shrubland (tall, mid,) Number of IBRA regions 53 Most extensive in IBRA region Est. pre-1750 and present : Great Victoria Desert (WA and SA) Estimated pre-1750 extent (km2) 865 845 Present extent (km2) 851 274 Area protected (km2) 85 444 Acacia ligulata (sandhill wattle), SA (Photo: M. -

Recommendation of Native Species for the Reforestation of Degraded Land Using Live Staking in Antioquia and Caldas’ Departments (Colombia)
UNIVERSITÀ DEGLI STUDI DI PADOVA Department of Land, Environment Agriculture and Forestry Second Cycle Degree (MSc) in Forest Science Recommendation of native species for the reforestation of degraded land using live staking in Antioquia and Caldas’ Departments (Colombia) Supervisor Prof. Lorenzo Marini Co-supervisor Prof. Jaime Polanía Vorenberg Submitted by Alicia Pardo Moy Student N. 1218558 2019/2020 Summary Although Colombia is one of the countries with the greatest biodiversity in the world, it has many degraded areas due to agricultural and mining practices that have been carried out in recent decades. The high Andean forests are especially vulnerable to this type of soil erosion. The corporate purpose of ‘Reforestadora El Guásimo S.A.S.’ is to use wood from its plantations, but it also follows the parameters of the Forest Stewardship Council (FSC). For this reason, it carries out reforestation activities and programs and, very particularly, it is interested in carrying out ecological restoration processes in some critical sites. The study area is located between 2000 and 2750 masl and is considered a low Andean humid forest (bmh-MB). The average annual precipitation rate is 2057 mm and the average temperature is around 11 ºC. The soil has a sandy loam texture with low pH, which limits the amount of nutrients it can absorb. FAO (2014) suggests that around 10 genera are enough for a proper restoration. After a bibliographic revision, the genera chosen were Alchornea, Billia, Ficus, Inga, Meriania, Miconia, Ocotea, Protium, Prunus, Psidium, Symplocos, Tibouchina, and Weinmannia. Two inventories from 2013 and 2019, helped to determine different biodiversity indexes to check the survival of different species and to suggest the adequate characteristics of the individuals for a successful vegetative stakes reforestation. -

Riverina Local Land Services TSR Vegetation Guide
Travelling Stock Reserves Vegetation Guide Riverina Local Land Services This project has been funded by NSW Environmental Trust Riverina Local Land Services Travelling Stock Reserve Vegetation Guide Prepared for NSW Local Land Services Report for: Local Land Services Prepared by: Ian Davidson, Regeneration Solutions Pty Ltd Date: February 2020 Funded by: NSW Environmental Trust This work draws heavily on material from the website of the Office of Environment and Heritage. The authors of this guide do not claim authorship, nor accept responsibility for, content drawn from this site. All photos were taken by Ian Davidson unless stated otherwise. Cover photo: Flax-lily flowering on Old Gunbar stock route Contents Vegetation in the Riverina region 1 Vegetation classes of the Riverina Local Land Services region 2 Southern Tableland Wet Sclerophyll Forests 3 Upper Riverina Dry Sclerophyll Forests 4 Western Slopes Dry Sclerophyll Forests 5 Western Slopes Grassy Woodlands 6 Floodplain Transition Woodlands 7 Riverine Sandhill Woodlands 8 Inland Riverine Forests 9 Inland Floodplain Woodlands 10 Inland Floodplain Shrublands 11 Inland Rocky Hill Woodlands 12 Riverine Plain Woodlands 13 Riverine Plain Grasslands 14 Riverine Chenopod Shrublands 15 Sand Plain Mallee Woodlands 16 Semi-arid Sand Plain Woodlands 17 NSW and EPBC (Commonwealth) Endangered Ecological Communities (EECs) of the Riverina region 18 NSW Endangered Ecological Communities 19 Commonwealth EPBC EECs 20 Site managed species 21 Recommended plant identification references 21 iv Riverina -

Albizia Chevalieri, And
学位論文 Studies on bioactive compounds from Cameroonian medicinal plants, Acacia albida and Albizia chevalieri, and Japanese endophytes Xylaria sp. (カメルーン産薬用植物 Acacia albida と Albizia chevalieri と日本で分離された植物内生菌類 Xylaria sp.の生理活性物質に関する研究) Abdou Tchoukoua Dedication To My late mom DJEBBA M. Odile i Acknowledgments Acknowledgments This thesis was successful with the support of a number of people to whom I am greatly indebted. I would like to express my sincere gratitude to my supervisors, Prof. Dr. Yoshihito Shiono (Yamagata University, Japan) and Prof. Ngadjui T. Bonaventure (University of Yaounde I, Cameroon) for the guidance, support, and motivation during all the time of my research in their labs. Their help will always be deeply appreciated. I would also like to thank the JSPS-Ronpaku Program for financial support in Japan. I wish to express my gratitude to Prof. M. Hashimoto, Prof. Y. Tokuji, Prof. K. Kimura, and Prof. T. Koseki, for their participation as judging committee. I wish to thank all the people with whom I have worked and contributed to the development of this work. Great thanks to Prof. Dr. Özgen Çaliskan for my sojourn in her lab, and to Dr. Ibrahim Horo in offering his expertise in isolation of saponins. I wish to thank Prof. K. Kimura for analyses of biological activities. I thank Drs Eunsang Kwon and Hiroyuki Momma for measurements of ESI-MS spectra. I am particularly grateful to Dr. T. Tabopda for his contribution to the success of this investigation. It has been a pleasure working with you. I thank the staff members of the Department of Organic Chemistry of the University of Yaounde I for my university education.