Antimicrobial and Cleaning Modes of Action of Peroxide and Reactive Oxygen Species for Combating Biofilms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sodium Percarbonate: a Versatile Oxidizing Reagent
SPOTLIGHT 2969 SYNLETT Sodium Percarbonate: A Versatile Spotlight 335 Oxidizing Reagent Compiled by Nadiya Koukabi This feature focuses on a re- Nadiya Koukabi was born in Shiraz, Iran. She received her B.Sc. agent chosen by a postgradu- (2000) in Chemistry from the Islamic Azad University Firouzabad ate, highlighting the uses and Branch and her M.Sc. in Organic Chemistry from the Bu-Ali Sina preparation of the reagent in University under the guidance of Prof. M. A. Zolfigol. She is pres- current research ently working toward her Ph.D. degree under the supervision of Prof. A. Khazaei and Prof. M. A. Zolfigol. Her research interests fo- cuse on the development of new reagents and catalysts in organic reactions. Faculty of Chemistry, Bu-Ali Sina University, Hamedan 6517838683, Iran E-mail: [email protected] Introduction available and furthermore, it is more risky to handle. Con- sequently, the ability of SPC to release oxidative species Sodium percarbonate (SPC, Na2CO3·1.5H2O) is an inor- in an organic medium has made it an useful reagent in or- ganic, inexpensive, environmentally friendly, stable, and ganic synthesis.8–11 easily handled reagent that has an excellent shelf life. The + Na H H H H – O O O O O name ‘sodium percarbonate’ does not reflect the structure O + O Na H O O or true nature of the material. Its erroneous name is due to + O Na – H H O 1,2 H successive confusions in its structure. The structure of – O O + H Na + Na – SPC (Figure 1) has been determined, the cohesion of the – O O O O O H – + O Na adduct being due to hydrogen bonding between carbonate O – – – O O 3,4 O O + ions and hydrogen peroxide molecules. -

ABSTRACT WANG, GUAN. Synthesis and Application of Bleach Activators
ABSTRACT WANG, GUAN. Synthesis and Application of Bleach Activators Containing Various Cationic Groups and PET Fabric Decolorization using Fenton’s Reagent. (Under the direction of Dr. David Hinks.) CBAs are quaternary ammonium salts (QAS) that are extensively used in various applications. Although most of the QAS are generally believed to be nontoxic to humans, they will harm aquatic life both animals and plants. CBAs are applied in textile bleaching process and the wastewater after bleaching contains chemicals including bleach activators. Even though wastewater contains CBAs will be treated in a wastewater treatment plant, the treated water still contain these chemicals. The study of acute toxicity and genotoxicity of CBAs to aquatic organisms was conducted in this study. Eight new bleach activators and two bench mark cationic bleach activators were investigated. New invented cationic bleach activator 3- PBBC is 86 times less toxic in the Daphnia sp. Immobilization Test, 18 times less toxic in Algae Toxicity assay and 10 times less mutagenic in the Salmonella/microsome microsuspension assay in comparison with the benchmark product (TBBC). This confirmed that replacing the cationic group in CBAs using low toxicity ammines can reduce the toxicity of CBAs to aquatic organisms. An effective low temperature and neutral pH bleaching system was developed using CBAs. A comparison life-cycle assessment for conventional and innovational bleaching system was conducted in this study. Their relative environmental performance was analyzed and compared. Based on data from industry, pre-published research, lab-scale experiments and the Ecoinvent database, the life-cycle-inventory (LCI) for both innovative and conventional bleaching process was developed. -

The Decomposition Kinetics of Peracetic Acid and Hydrogen Peroxide in Municipal Wastewaters
Disinfection Forum No 10, October 2015 The Decomposition Kinetics of Peracetic Acid and Hydrogen Peroxide in Municipal Wastewaters INTRODUCTION Efficient control of microbial populations in municipal wastewater using peracetic acid (PAA) requires an understanding of the PAA decomposition kinetics. This knowledge is critical to ensure the proper dosing of PAA needed to achieve an adequate concentration within the contact time of the disinfection chamber. In addition, the impact of PAA on the environment, post-discharge into the receiving water body, also is dependent upon the longevity of the PAA in the environment, before decomposing to acetic acid, oxygen and water. As a result, the decomposition kinetics of PAA may have a significant impact on aquatic and environmental toxicity. PAA is not manufactured as a pure compound. The solution exists as an equilibrium mixture of PAA, hydrogen peroxide, acetic acid, and water: ↔ + + Acetic Acid Hydrogen Peroxide Peracetic Acid Water PeroxyChem’s VigorOx® WWT II Wastewater Disinfection Technology contains 15% peracetic acid by weight and 23% hydrogen peroxide as delivered. Although hydrogen peroxide is present in the formulation, peracetic acid is considered to be the active component for disinfection1 in wastewater. There have been several published studies investigating the decomposition kinetics of PAA in different water matrices, including municipal wastewater2-7. Yuan7 states that PAA may be consumed in the following three competitive reactions: 1. Spontaneous decomposition 2 CH3CO3H à 2 CH3CO2H + O2 Eq (1) 2. Hydrolysis CH3CO3H + H2O à CH3CO2H + H2O2 Eq (2) 3. Transition metal catalyzed decomposition + CH3CO3H + M à CH3CO2H + O2 + other products Eq (3) At neutral pH’s, both peracetic acid and hydrogen peroxide can be rapidly consumed by these reactions7 (hydrogen peroxide will decompose to water and oxygen via 2H2O2 à 2H2O + O2). -

Peracetic Acid Processing
Peracetic Acid Processing Identification Chemical Name(s): CAS Number: peroxyacetic acid, ethaneperoxic acid 79-21-0 Other Names: Other Codes: per acid, periacetic acid, PAA NIOSH Registry Number: SD8750000 TRI Chemical ID: 000079210 UN/ID Number: UN3105 Summary Recommendation Synthetic / Allowed or Suggested Non-Synthetic: Prohibited: Annotation: Synthetic Allowed (consensus) Allowed only for direct food contact for use in wash water. Allowed as a (consensus) sanitizer on surfaces in contact with organic food. (consensus) From hydrogen peroxide and fermented acetic acid sources only. (Not discussed by processing reviewers--see discussion of source under Crops PAA TAP review.) Characterization Composition: C2H4O3. Peracetic acid is a mixture of acetic acid (CH3COOH) and hydrogen peroxide (H2O2) in an aqueous solution. Acetic acid is the principle component of vinegar. Hydrogen peroxide has been previously recommended by the NOSB for the National List in processing (synthetic, allowed at Austin, 1995). Properties: It is a very strong oxidizing agent and has stronger oxidation potential than chlorine or chlorine dioxide. Liquid, clear, and colorless with no foaming capability. It has a strong pungent acetic acid odor, and the pH is acid (2.8). Specific gravity is 1.114 and weighs 9.28 pounds per gallon. Stable upon transport. How Made: Peracetic acid (PAA) is produced by reacting acetic acid and hydrogen peroxide. The reaction is allowed to continue for up to ten days in order to achieve high yields of product according to the following equation. O O || || CH3-C-OH + H2O2 CH3C-O-OH + H2O acetic acid hydrogen peroxyacetic peroxide acid Due to reaction limitations, PAA generation can be up to 15% with residual levels of hydrogen peroxide (up to 25%) and acetic acid (up to 35%) with water up to 25%. -

Great People Great Products Perfect Chemistry Oci
OCI CHEMICAL CORPORATION GREAT PEOPLE GREAT PRODUCTS PERFECT CHEMISTRY OCI Chemical is a subsidiary of OCI Company Ltd., a leading chemical manufacturer and publicly traded company on the Korean Exchange. OCI Company has five primary business divisions: renewable energy, inorganic chemicals, petro and coal chemicals, fine chemicals and insulation. OCI Chemical is an integral part of the inorganic chemicals division and a major contributor to this division’s sales and profits. Since 1962, OCI Chemical’s plant in Green River, Wyoming has been providing high quality soda ash to companies across North America and around the globe. While we are still among the world’s largest producers of soda ash, OCI Chemical has also become a leading producer of sodium percarbonate (PC) and hydrogen peroxide. A GLOBAL LEADER A GLOBAL OCI Chemical is committed to excellence and safety in all aspects of business. Every OCI employee, from the plants in Green River, Decatur and Columbus to our corporate headquarters in Atlanta, is empowered to modify any operation that poses a potential threat to themselves, their fellow workers, the surrounding community or the environment. Our customers know they can count on us for more than just quality chemicals. They know OCI was built on strong values, strong character and a passion for excellence. We know that great people and great products equal perfect chemistry. OCI Wyoming, L.P. Green River, Wyoming Soda Ash Mining and Production Stauffer Chemical opens Big THROUGH THE YEARS THROUGH THE Island Mine and Refinery in Green River, WY and produces 1962 soda ash from the mined trona. -

Peracetic Acid
Regulation (EU) No 528/2012 concerning the making available on the market and use of biocidal products Evaluation of active substances Assessment Report Peracetic acid Product-types 11 and 12 (Preservatives for liquid cooling and processing systems) (Slimicides) August 2016 Finland Peracetic acid Product-types 11 and 12 August 2016 CONTENTS 1. STATEMENT OF SUBJECT MATTER AND PURPOSE ........................................................ 2 1.1. Procedure followed ............................................................................................. 2 1.2. Purpose of the assessment report ....................................................................... 2 2. OVERALL SUMMARY AND CONCLUSIONS ..................................................................... 4 2.1. Presentation of the Active Substance .................................................................. 4 2.1.1. Identity, Physico-Chemical Properties & Methods of Analysis ................. 4 2.1.2. Intended Uses and Efficacy ..................................................................... 7 2.1.3. Classification and Labelling ..................................................................... 8 2.2. Summary of the Risk Assessment ....................................................................... 9 2.2.1. Human Health Risk Assessment .............................................................. 9 2.2.1.1. Hazard identification ................................................................................... 9 2.2.1.2. Effects assessment ................................................................................... -
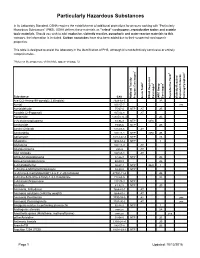
Particularly Hazardous Substances
Particularly Hazardous Substances In its Laboratory Standard, OSHA requires the establishment of additional protections for persons working with "Particularly Hazardous Substances" (PHS). OSHA defines these materials as "select" carcinogens, reproductive toxins and acutely toxic materials. Should you wish to add: explosive, violently reactive, pyrophoric and water-reactve materials to this category, the information is included. Carbon nanotubes have also been added due to their suspected carcinogenic properties. This table is designed to assist the laboratory in the identification of PHS, although it is not definitively conclusive or entirely comprehensive. *Notes on the proper use of this table appear on page 12. 1 6 5 2 3 4 Substance CAS National Toxicity National Program Carcinogen Toxin Acute Regulated OSHA Carcinogen Group IARC Carcinogen Toxin Reproductive Violently Reactive/ Explosive/Peroxide Forming/Pyrophoric A-a-C(2-Amino-9H-pyrido[2,3,b]indole) 2648-68-5 2B Acetal 105-57-7 yes Acetaldehyde 75-07-0 NTP AT 2B Acrolein (2-Propenal) 107-02-8 AT Acetamide 126850-14-4 2B 2-Acetylaminofluorene 53-96-3 NTP ORC Acrylamide 79-06-6 NTP 2B Acrylyl Chloride 814-68-6 AT Acrylonitrile 107-13-1 NTP ORC 2B Adriamycin 23214-92-8 NTP 2A Aflatoxins 1402-68-2 NTP 1 Allylamine 107-11-9 AT Alkylaluminums varies AT Allyl Chloride 107-05-1 AT ortho-Aminoazotoluene 97-56-3 NTP 2B para-aminoazobenzene 60-09-3 2B 4-Aminobiphenyl 92-67-1 NTP ORC 1 1-Amino-2-Methylanthraquinone 82-28-0 NTP (2-Amino-6-methyldipyrido[1,2-a:3’,2’-d]imidazole) 67730-11-4 2B -

UCLA Electronic Theses and Dissertations
UCLA UCLA Electronic Theses and Dissertations Title Use of Boron in Detergents and its Impact on Reclamation Permalink https://escholarship.org/uc/item/2rw7k2r7 Author Ghavanloughajar, Maryam Publication Date 2015 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Use of Boron in Detergents and its Impact on Reclamation A thesis submitted in partial satisfaction of the requirements for the degree Master of Science in Civil Engineering by Maryam Ghavanloughajar 2015 ABSTRACT OF THE THESIS Use of Boron in Detergents and its Impact on Reclamation By Maryam Ghavanloughajar Master of Science in Civil Engineering University of California, Los Angeles, 2015 Professor Michael K. Stenstrom, Chair Many parts of the world are experiencing severe water drought and it affects societies both economically and environmentally. Therefore, conservation practices are essential to balance water supply and demand. Greywater or wastewaters from showers and luandries, if treated well can be a reliable source for activities such as irrigation, toilet flushing and car washing. Greywaters are not as contaminated as sewage but still may require treatment before reuse. The application of insufficiently treated water for irrigation can cause harm to plants and animals. Pollutant such as boron in greywater is of particular interest because many plants are sensitive to even low concentrations. High concentrations of boron can induce toxicity, reduce growth rate and yield in plants. Therefore, proposed greywater treatment systems need to consider the sensitivity of plant species and boron concentrations and potential removal. This thesis reviews boron chemistry, its effect on plants and currently available boron removal technologies. -

Peracetic Acid Bleaching CH CO H
Peracetic Acid Bleaching CH3CO3H Introduction of Bleaching Bleaching is a chemical decoloration and delignification process carried out on various types of pulp. DliDelignifi cati on Removal of the chromophores At in dus tr ia l scal e, bl eachi ng i s perf ormed b y chl ori ne, chl ori ne dioxide, oxygen, hydrogen peroxide, ozone and peracetic acid. Effic iency: ClO2 = Cl2 = O3 >CH3COOOH > H2O2 >O> O2 Often bleaching chemicals are wasted in secondary reactions. D. Lachenal and C. Chirat, Cellulose Chem. Technol., 39, 5-6, 511-156 (2005) 2 Electron Exchange Theory Ring opening of the aromatic units necessitates the exchange of 4 e - # of e- can be exchanged per bleaching molecule - + O3 + 6 e + 6 H → 3 H2O - + - CCOlO2 + 55e e + 4 H → Cl + 2 H2O2 - + O2 + 4 e + 4 H → 2 H2O - + H2O2 + 2 e + 2 H → 2 H2O - + CH3COOOH + 2 e + 2 H → CH3COOH + H2O # of e- exchanged per molecule decreases in the following order: O3 > ClO2 > O2 > H2O2 = CH3COOOH D. Lachenal and C. Chirat, Cellulose Chem. Technol., 39, 5-6, 511-156 (2005) 3 Actual Electron Exchange - ClO2 and O3 100 g un bleac he d SW k raf t pul p, k appa # f rom 30 to ~ 4 -5 - 2.3% ClO2 (0.034 mole) on pulp: 0.034 x 5 = 0.17 e is exchanged - 14%1.4% O3 (0.03 mole)on pulp: 0 .03 x 6 = 018e0.18 e is exchanged Chemicals % consumed1 Kappa number * ClO2 2.3 4.5 O3 1.5 3.5 O3 1.4 5.0 O3 1.3 6.5 1Chemicals consumed are shown as % on pulp. -
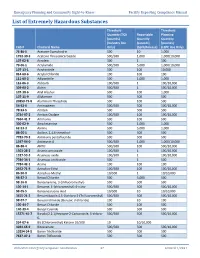
List of Extremely Hazardous Substances
Emergency Planning and Community Right-to-Know Facility Reporting Compliance Manual List of Extremely Hazardous Substances Threshold Threshold Quantity (TQ) Reportable Planning (pounds) Quantity Quantity (Industry Use (pounds) (pounds) CAS # Chemical Name Only) (Spill/Release) (LEPC Use Only) 75-86-5 Acetone Cyanohydrin 500 10 1,000 1752-30-3 Acetone Thiosemicarbazide 500/500 1,000 1,000/10,000 107-02-8 Acrolein 500 1 500 79-06-1 Acrylamide 500/500 5,000 1,000/10,000 107-13-1 Acrylonitrile 500 100 10,000 814-68-6 Acrylyl Chloride 100 100 100 111-69-3 Adiponitrile 500 1,000 1,000 116-06-3 Aldicarb 100/500 1 100/10,000 309-00-2 Aldrin 500/500 1 500/10,000 107-18-6 Allyl Alcohol 500 100 1,000 107-11-9 Allylamine 500 500 500 20859-73-8 Aluminum Phosphide 500 100 500 54-62-6 Aminopterin 500/500 500 500/10,000 78-53-5 Amiton 500 500 500 3734-97-2 Amiton Oxalate 100/500 100 100/10,000 7664-41-7 Ammonia 500 100 500 300-62-9 Amphetamine 500 1,000 1,000 62-53-3 Aniline 500 5,000 1,000 88-05-1 Aniline, 2,4,6-trimethyl- 500 500 500 7783-70-2 Antimony pentafluoride 500 500 500 1397-94-0 Antimycin A 500/500 1,000 1,000/10,000 86-88-4 ANTU 500/500 100 500/10,000 1303-28-2 Arsenic pentoxide 100/500 1 100/10,000 1327-53-3 Arsenous oxide 100/500 1 100/10,000 7784-34-1 Arsenous trichloride 500 1 500 7784-42-1 Arsine 100 100 100 2642-71-9 Azinphos-Ethyl 100/500 100 100/10,000 86-50-0 Azinphos-Methyl 10/500 1 10/10,000 98-87-3 Benzal Chloride 500 5,000 500 98-16-8 Benzenamine, 3-(trifluoromethyl)- 500 500 500 100-14-1 Benzene, 1-(chloromethyl)-4-nitro- 500/500 -

Sodium Percarbonate
Technical Data Sheet for OCI Sodium Percarbonate Sodium Percarbonate A Guide for High Performance Eco-Friendly Bleaching Version 1.0 at May 7th, 2009 by Houngshup. Shin, HERO Team, Iksan Plant, OCI 1. Introduction 1.1. OCI – Company profile Welcome to OCI. We would like to extend our sincere appreciation to all who have continuously supported OCI. Since being founded in 1959, as a leading company in the chemical industry, we have dedicated ourselves solely to the enhancement of the chemical industry. Today, OCI has grown into a leader in the fields of basic chemical, fine chemical, and petrochemical, as well as coal chemical and material processing industry. Based on our technology and outstanding personnel, we promise to build an affluent and environment-friendly society for all our customers. OCI is one of the world’s biggest leading sodium percarbonate producers, with a capacity of 180,000MT/year and plants in Korea (OCI Iksan Plant), China (Zhejiang OCI) and USA (OCI Chemical Corporation). OCI acquired a plant in Zhejiang, China from Shangyu Jiehua Chemical in 2006 and established OCI Chemical Corporation in the United States in 2001. As the leading supplier of sodium percarbonate, OCI worldwidely offers not only the full range of sodium percarbonate but also technical services through its global network. 1.2. Sodium percarbonate (Synonyms: sodium carbonate peroxyhydrate, sodium carbonate peroxide, carbonic acid disodium salt compound with hydrogen peroxide. Abbrev.: SPC, PCS etc., CAS No. 15630- 89-4) is an adduct of hydrogen peroxide and sodium carbonate (soda ash). Based on the molecular formula, the pure substance sodium percarbonate contains 32.5% hydrogen peroxide and 67.5% sodium carbonate (based on weight). -

(12) Patent Application Publication (10) Pub. No.: US 2016/0372276 A1 HAN Et Al
US 20160372276A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0372276 A1 HAN et al. (43) Pub. Date: Dec. 22, 2016 (54) A PRECIOUS METAL SWITCH CONTACT C2.5D 5/48 (2006.01) COMPONENT AND ITS PREPARATION C2.5D 3/48 (2006.01) METHOD C2.5D 3/46 (2006.01) C2.5D 5/02 (2006.01) (71) Applicant: NANTONG MEMTECH HIH II/04 (2006.01) TECHNOLOGIES CO., LTD, C23C 8/42 (2006.01) Nantong (CN) (52) U.S. Cl. CPC ................ H01H I/02 (2013.01); H0IH II/04 (72) Inventors: Huisheng HAN, Nantong (CN); (2013.01); C23C 18/1605 (2013.01); C23C HONGMEI ZHANG, NANTONG 18/1689 (2013.01); C23C 18/42 (2013.01); (CN); YANG DING, NANTONG (CN); C25D 3/48 (2013.01); C25D 3/46 (2013.01); ZHIHONG DONG, NANTONG (CN); C25D 5/022 (2013.01); C25D 5/48 (2013.01) CHENG HUANG, NANTONG (CN) (57) ABSTRACT (21) Appl. No.: 14/896,403 This invention discloses a preparation method for precious metal Switching contact components by means of plating (22) PCT Filed: Sep. 15, 2014 masking, plating and etching processes. The plating masking PCT/CN2014/090913 process is performed by using a plating mask ink with or (86). PCT No.: without a photo exposure machine. Plating of precious S 371 (c)(1), metals is performed by electroless plating or electro plating (2) Date: Dec. 7, 2015 methods. Etching is carried out with etching solutions con taining weak organic acids, weak inorganic acids or acidic (30) Foreign Application Priority Data buffering agents. Improvement of the etched Surface gloss and prevention of the side etching are realized with the Sep.