DNA Repair - Takahiko Taguchi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolutionary Origins of DNA Repair Pathways: Role of Oxygen Catastrophe in the Emergence of DNA Glycosylases
cells Review Evolutionary Origins of DNA Repair Pathways: Role of Oxygen Catastrophe in the Emergence of DNA Glycosylases Paulina Prorok 1 , Inga R. Grin 2,3, Bakhyt T. Matkarimov 4, Alexander A. Ishchenko 5 , Jacques Laval 5, Dmitry O. Zharkov 2,3,* and Murat Saparbaev 5,* 1 Department of Biology, Technical University of Darmstadt, 64287 Darmstadt, Germany; [email protected] 2 SB RAS Institute of Chemical Biology and Fundamental Medicine, 8 Lavrentieva Ave., 630090 Novosibirsk, Russia; [email protected] 3 Center for Advanced Biomedical Research, Department of Natural Sciences, Novosibirsk State University, 2 Pirogova St., 630090 Novosibirsk, Russia 4 National Laboratory Astana, Nazarbayev University, Nur-Sultan 010000, Kazakhstan; [email protected] 5 Groupe «Mechanisms of DNA Repair and Carcinogenesis», Equipe Labellisée LIGUE 2016, CNRS UMR9019, Université Paris-Saclay, Gustave Roussy Cancer Campus, F-94805 Villejuif, France; [email protected] (A.A.I.); [email protected] (J.L.) * Correspondence: [email protected] (D.O.Z.); [email protected] (M.S.); Tel.: +7-(383)-3635187 (D.O.Z.); +33-(1)-42115404 (M.S.) Abstract: It was proposed that the last universal common ancestor (LUCA) evolved under high temperatures in an oxygen-free environment, similar to those found in deep-sea vents and on volcanic slopes. Therefore, spontaneous DNA decay, such as base loss and cytosine deamination, was the Citation: Prorok, P.; Grin, I.R.; major factor affecting LUCA’s genome integrity. Cosmic radiation due to Earth’s weak magnetic field Matkarimov, B.T.; Ishchenko, A.A.; and alkylating metabolic radicals added to these threats. -

Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA
Mechanisms in E. coli and Human Mismatch Repair Nobel Lecture, December 8, 2015 by Paul Modrich Howard Hughes Medical Institute and Department of Biochemistry, Duke University Medical Center, Durham, North Carolina, USA. he idea that mismatched base pairs occur in cells and that such lesions trig- T ger their own repair was suggested 50 years ago by Robin Holliday in the context of genetic recombination [1]. Breakage and rejoining of DNA helices was known to occur during this process [2], with precision of rejoining attributed to formation of a heteroduplex joint, a region of helix where the two strands are derived from the diferent recombining partners. Holliday pointed out that if this heteroduplex region should span a genetic diference between the two DNAs, then it will contain one or more mismatched base pairs. He invoked processing of such mismatches to explain the recombination-associated phenomenon of gene conversion [1], noting that “If there are enzymes which can repair points of damage in DNA, it would seem possible that the same enzymes could recognize the abnormality of base pairing, and by exchange reactions rectify this.” Direct evidence that mismatches provoke a repair reaction was provided by bacterial transformation experiments [3–5], and our interest in this efect was prompted by the Escherichia coli (E. coli) work done in Matt Meselson’s lab at Harvard. Using artifcially constructed heteroduplex DNAs containing multiple mismatched base pairs, Wagner and Meselson [6] demonstrated that mismatches elicit a repair reaction upon introduction into the E. coli cell. Tey also showed that closely spaced mismatches, mismatches separated by a 1000 base pairs or so, are usually repaired on the same DNA strand. -

Lecture: 28 TRANSAMINATION, DEAMINATION and DECARBOXYLATION
Lecture: 28 TRANSAMINATION, DEAMINATION AND DECARBOXYLATION Protein metabolism is a key physiological process in all forms of life. Proteins are converted to amino acids and then catabolised. The complete hydrolysis of a polypeptide requires mixture of peptidases because individual peptidases do not cleave all peptide bonds. Both exopeptidases and endopeptidases are required for complete conversion of protein to amino acids. Amino acid metabolism The amino acids not only function as energy metabolites but also used as precursors of many physiologically important compounds such as heme, bioactive amines, small peptides, nucleotides and nucleotide coenzymes. In normal human beings about 90% of the energy requirement is met by oxidation of carbohydrates and fats. The remaining 10% comes from oxidation of the carbon skeleton of amino acids. Since the 20 common protein amino acids are distinctive in terms of their carbon skeletons, amino acids require unique degradative pathway. The degradation of the carbon skeletons of 20 amino acids converges to just seven metabolic intermediates namely. i. Pyruvate ii. Acetyl CoA iii. Acetoacetyl CoA iv. -Ketoglutarate v. Succinyl CoA vi. Fumarate vii. Oxaloacetate Pyruvate, -ketoglutarate, succinyl CoA, fumarate and oxaloacetate can serve as precursors for glucose synthesis through gluconeogenesis.Amino acids giving rise to these intermediates are termed as glucogenic. Those amino acids degraded to yield acetyl CoA or acetoacetate are termed ketogenic since these compounds are used to synthesize ketone bodies. Some amino acids are both glucogenic and ketogenic (For example, phenylalanine, tyrosine, tryptophan and threonine. Catabolism of amino acids The important reaction commonly employed in the breakdown of an amino acid is always the removal of its -amino group. -

DNA Repair with Its Consequences (E.G
Cell Science at a Glance 515 DNA repair with its consequences (e.g. tolerance and pathways each require a number of apoptosis) as well as direct correction of proteins. By contrast, O-alkylated bases, Oliver Fleck* and Olaf Nielsen* the damage by DNA repair mechanisms, such as O6-methylguanine can be Department of Genetics, Institute of Molecular which may require activation of repaired by the action of a single protein, Biology, University of Copenhagen, Øster checkpoint pathways. There are various O6-methylguanine-DNA Farimagsgade 2A, DK-1353 Copenhagen K, Denmark forms of DNA damage, such as base methyltransferase (MGMT). MGMT *Authors for correspondence (e-mail: modifications, strand breaks, crosslinks removes the alkyl group in a suicide fl[email protected]; [email protected]) and mismatches. There are also reaction by transfer to one of its cysteine numerous DNA repair pathways. Each residues. Photolyases are able to split Journal of Cell Science 117, 515-517 repair pathway is directed to specific Published by The Company of Biologists 2004 covalent bonds of pyrimidine dimers doi:10.1242/jcs.00952 types of damage, and a given type of produced by UV radiation. They bind to damage can be targeted by several a UV lesion in a light-independent Organisms are permanently exposed to pathways. Major DNA repair pathways process, but require light (350-450 nm) endogenous and exogenous agents that are mismatch repair (MMR), nucleotide as an energy source for repair. Another damage DNA. If not repaired, such excision repair (NER), base excision NER-independent pathway that can damage can result in mutations, diseases repair (BER), homologous recombi- remove UV-induced damage, UVER, is and cell death. -

DNA DEAMINATION REPAIR ENZYMES in BACTERIAL and HUMAN SYSTEMS Rongjuan Mi Clemson University, [email protected]
Clemson University TigerPrints All Dissertations Dissertations 12-2008 DNA DEAMINATION REPAIR ENZYMES IN BACTERIAL AND HUMAN SYSTEMS Rongjuan Mi Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_dissertations Part of the Biochemistry Commons Recommended Citation Mi, Rongjuan, "DNA DEAMINATION REPAIR ENZYMES IN BACTERIAL AND HUMAN SYSTEMS" (2008). All Dissertations. 315. https://tigerprints.clemson.edu/all_dissertations/315 This Dissertation is brought to you for free and open access by the Dissertations at TigerPrints. It has been accepted for inclusion in All Dissertations by an authorized administrator of TigerPrints. For more information, please contact [email protected]. DNA DEAMINATION REPAIR ENZYMES IN BACTERIAL AND HUMAN SYSTEMS A Dissertation Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Biochemistry by Rongjuan Mi December 2008 Accepted by: Dr. Weiguo Cao, Committee Chair Dr. Chin-Fu Chen Dr. James C. Morris Dr. Gary Powell ABSTRACT DNA repair enzymes and pathways are diverse and critical for living cells to maintain correct genetic information. Single-strand-selective monofunctional uracil DNA glycosylase (SMUG1) belongs to Family 3 of the uracil DNA glycosylase superfamily. We report that a bacterial SMUG1 ortholog in Geobacter metallireducens (Gme) and the human SMUG1 enzyme are not only uracil DNA glycosylases (UDG), but also xanthine DNA glycosylases (XDG). Mutations at M57 (M57L) and H210 (H210G, H210M, H210N) can cause substantial reductions in XDG and UDG activities. Increased selectivity is achieved in the A214R mutant of Gme SMUG1 and G60Y completely abolishes XDG and UDG activity. Most interestingly, a proline substitution at the G63 position switches the Gme SMUG1 enzyme to an exclusive uracil DNA glycosylase. -

DNA Proofreading and Repair
DNA proofreading and repair Mechanisms to correct errors during DNA replication and to repair DNA damage over the cell's lifetime. Key points: Cells have a variety of mechanisms to prevent mutations, or permanent changes in DNA sequence. During DNA synthesis, most DNA polymerases "check their work," fixing the majority of mispaired bases in a process called proofreading. Immediately after DNA synthesis, any remaining mispaired bases can be detected and replaced in a process called mismatch repair. If DNA gets damaged, it can be repaired by various mechanisms, including chemical reversal, excision repair, and double-stranded break repair. Introduction What does DNA have to do with cancer? Cancer occurs when cells divide in an uncontrolled way, ignoring normal "stop" signals and producing a tumor. This bad behavior is caused by accumulated mutations, or permanent sequence changes in the cells' DNA. Replication errors and DNA damage are actually happening in the cells of our bodies all the time. In most cases, however, they don’t cause cancer, or even mutations. That’s because they are usually detected and fixed by DNA proofreading and repair mechanisms. Or, if the damage cannot be fixed, the cell will undergo programmed cell death (apoptosis) to avoid passing on the faulty DNA. Mutations happen, and get passed on to daughter cells, only when these mechanisms fail. Cancer, in turn, develops only when multiple mutations in division-related genes accumulate in the same cell. In this article, we’ll take a closer look at the mechanisms used by cells to correct replication errors and fix DNA damage, including: Proofreading, which corrects errors during DNA replication Mismatch repair, which fixes mispaired bases right after DNA replication DNA damage repair pathways, which detect and correct damage throughout the cell cycle Proofreading DNA polymerases are the enzymes that build DNA in cells. -

Clinical Significance of ERCC2, XPC, ERCC5 and XRCC3 Gene Polymorphisms in Diffuse Large B Cell Lymphoma
DOI: 10.14744/ejmi.2020.56831 EJMI 2020;4(3):332–340 Research Article Clinical Significance of ERCC2, XPC, ERCC5 and XRCC3 Gene Polymorphisms in Diffuse Large B Cell Lymphoma Aykut Bahceci,1 Semra Paydas,2 Melek Ergin,3 Gulsah Seydaoglu,4 Gulsum Ucar5 1Department of Medical Oncology, Dr. Ersin Arslan Training and Research Hospital, Gaziantep, Turkey 2Department of Medical Oncology, Cukurova University Faculty of Medicine, Adana, Turkey 3Department of Patology, Cukurova University Faculty of Medicine, Adana, Turkey 4Department of Biostatistics, Cukurova University Faculty of Medicine, Adana, Turkey 5Department of Pediatric Hematology, Cukurova University Faculty of Medicine, Adana, Turkey Abstract Objectives: DNA repair genes protects the genome from DNA damage both of endogenous and exogenous stress fac- tors. Due to DNA repair gene polymorphisms, there are differences in the repair capacity between several cancer types. The aim of this study is to evaluate the association between some of the DNA repair gene polymorphisms and clinical outcome in Diffuse Large B-Cell Lymphoma (DLBCL). Methods: The association between clinical factors including stage at diagnosis, extra-nodal involvement, tumor bur- den, bone marrow involvement, relapse status, disease-free/overall survival times and DNA repair gene polymorphisms including ERCC2 (Lys751Gln), XPC (Gln939Lys), ERCC5 (Asp1104His) and XRCC3 (Thr241Met) in 58 patients with DLBCL. T-Shift Real-Time PCR was used to detect these mutations. Results: The median survival times were 60 months and 109 months in patients with CC genotype and CA/AA geno- type of XPC gene polymorphism, respectively (p=0.017). More interestingly, median survival times were 9 months and 109 months in patients with CC (XPC)/CC (XRCC3) and CA/AA (XPC)/CT/TT (XRCC3) for both XPC and XRCC3 gene polymorphisms, respectively (p=0.004). -

Predisposition to Hematologic Malignancies in Patients With
LETTERS TO THE EDITOR carcinomas but no internal cancer by the age of 29 years Predisposition to hematologic malignancies in and 9 years, respectively. patients with xeroderma pigmentosum Case XP540BE . This patient had a highly unusual pres - entation of MPAL. She was diagnosed with XP at the age Germline predisposition is a contributing etiology of of 18 months with numerous lentigines on sun-exposed hematologic malignancies, especially in children and skin, when her family emigrated from Morocco to the young adults. Germline predisposition in myeloid neo - USA. The homozygous North African XPC founder muta - plasms was added to the World Health Organization tion was present. 10 She had her first skin cancer at the age 1 2016 classification, and current management recommen - of 8 years, and subsequently developed more than 40 cuta - dations emphasize the importance of screening appropri - neous basal and squamous cell carcinomas, one melanoma 2 ate patients. Rare syndromes of DNA repair defects can in situ , and one ocular surface squamous neoplasm. She 3 lead to myeloid and/or lymphoid neoplasms. Here, we was diagnosed with a multinodular goiter at the age of 9 describe our experience with hematologic neoplasms in years eight months, with several complex nodules leading the defective DNA repair syndrome, xeroderma pigmen - to removal of her thyroid gland. Histopathology showed tosum (XP), including myelodysplastic syndrome (MDS), multinodular adenomatous/papillary hyperplasia. At the secondary acute myeloid leukemia (AML), high-grade age of 19 years, she presented with night sweats, fatigue, lymphoma, and an extremely unusual presentation of and lymphadenopathy. Laboratory studies revealed pancy - mixed phenotype acute leukemia (MPAL) with B, T and topenia with hemoglobin 6.8 g/dL, platelet count myeloid blasts. -

The Consequences of Rad51 Overexpression for Normal and Tumor Cells
dna repair 7 (2008) 686–693 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/dnarepair Mini review The consequences of Rad51 overexpression for normal and tumor cells Hannah L. Klein ∗ Department of Biochemistry, New York University School of Medicine, NYU Medical Center, 550 First Avenue, New York, NY 10016, United States article info abstract Article history: The Rad51 recombinase is an essential factor for homologous recombination and the Received 11 December 2007 repair of DNA double strand breaks, binding transiently to both single stranded and double Accepted 12 December 2007 stranded DNA during the recombination reaction. The use of a homologous recombination Published on line 1 February 2008 mechanism to repair DNA damage is controlled at several levels, including the binding of Rad51 to single stranded DNA to form the Rad51 nucleofilament, which is controlled through Keywords: the action of DNA helicases that can counteract nucleofilament formation. Overexpression Rad51 protein of Rad51 in different organisms and cell types has a wide assortment of consequences, rang- Overexpression of Rad51 ing from increased homologous recombination and increased resistance to DNA damaging Genomic instability agents to disruption of the cell cycle and apoptotic cell death. Rad51 expression is increased Tumor cell drug resistance in p53-negative cells, and since p53 is often mutated in tumor cells, there is a tendency for Homologous recombination Rad51 to be overexpressed in tumor cells, leading to increased resistance to DNA damage Gene targeting and drugs used in chemotherapies. As cells with increased Rad51 levels are more resis- tant to DNA damage, there is a selection for tumor cells to have higher Rad51 levels. -

Amino Acid Catabolism: Urea Cycle the Urea Bi-Cycle Two Issues
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Urea Cycle – dealing with the nitrogen Protein turnover Ubiquitin Feeding the Urea Cycle Activation-E1 Glucose-Alanine Cycle Conjugation-E2 Free Ammonia Ligation-E3 Proteosome Glutamine Amino-Acid Degradation Glutamate dehydrogenase Ammonia Overall energetics free Dealing with the carbon transamination-mechanism to know Seven Families Urea Cycle – dealing with the nitrogen 1. ADENQ 5 Steps 2. RPH Carbamoyl-phosphate synthetase oxidase Ornithine transcarbamylase one-carbon metabolism Arginino-succinate synthetase THF Arginino-succinase SAM Arginase 3. GSC Energetics PLP uses Urea Bi-cycle 4. MT – one carbon metabolism 5. FY – oxidases Amino Acid Catabolism: Urea Cycle The Urea Bi-Cycle Two issues: 1) What to do with the fumarate? 2) What are the sources of the free ammonia? a-ketoglutarate a-amino acid Aspartate transaminase transaminase a-keto acid Glutamate 1 Amino Acid Catabolism: Urea Cycle The Glucose-Alanine Cycle • Vigorously working muscles operate nearly anaerobically and rely on glycolysis for energy. a-Keto acids • Glycolysis yields pyruvate. – If not eliminated (converted to acetyl- CoA), lactic acid will build up. • If amino acids have become a fuel source, this lactate is converted back to pyruvate, then converted to alanine for transport into the liver. Excess Glutamate is Metabolized in the Mitochondria of Hepatocytes Amino Acid Catabolism: Urea Cycle Excess glutamine is processed in the intestines, kidneys, and liver. (deaminating) (N,Q,H,S,T,G,M,W) OAA à Asp Glutamine Synthetase This costs another ATP, bringing it closer to 5 (N,Q,H,S,T,G,M,W) 29 N 2 Amino Acid Catabolism: Urea Cycle Excess glutamine is processed in the intestines, kidneys, and liver. -
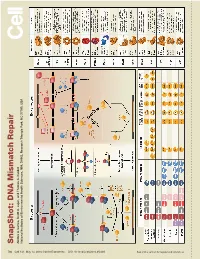
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

Involvement of Nucleotide Excision and Mismatch Repair Mechanisms in Double Strand Break Repair
250 Current Genomics, 2009, 10, 250-258 Involvement of Nucleotide Excision and Mismatch Repair Mechanisms in Double Strand Break Repair Ye Zhang1,2,*, Larry H. Rohde2 and Honglu Wu1 1NASA Johnson Space Center, Houston, Texas 77058 and 2University of Houston at Clear Lake, Houston, Texas 77058, USA Abstract: Living organisms are constantly threatened by environmental DNA-damaging agents, including UV and ioniz- ing radiation (IR). Repair of various forms of DNA damage caused by IR is normally thought to follow lesion-specific re- pair pathways with distinct enzymatic machinery. DNA double strand break is one of the most serious kinds of damage induced by IR, which is repaired through double strand break (DSB) repair mechanisms, including homologous recombi- nation (HR) and non-homologous end joining (NHEJ). However, recent studies have presented increasing evidence that various DNA repair pathways are not separated, but well interlinked. It has been suggested that non-DSB repair mecha- nisms, such as Nucleotide Excision Repair (NER), Mismatch Repair (MMR) and cell cycle regulation, are highly involved in DSB repairs. These findings revealed previously unrecognized roles of various non-DSB repair genes and indicated that a successful DSB repair requires both DSB repair mechanisms and non-DSB repair systems. One of our recent studies found that suppressed expression of non-DSB repair genes, such as XPA, RPA and MLH1, influenced the yield of IR- induced micronuclei formation and/or chromosome aberrations, suggesting that these genes are highly involved in DSB repair and DSB-related cell cycle arrest, which reveals new roles for these gene products in the DNA repair network.