Labelling Accuracy in Tasmanian Seafood: an Investigation Using DNA Barcoding
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

OVERVIEW of FOOD FRAUD in the FISHERIES SECTOR Cover Photo: Mussel Farm in the Philippines
FIAM/C1165 (En) FAO Fisheries and Aquaculture Circular ISSN 2070-6065 OVERVIEW OF FOOD FRAUD IN THE FISHERIES SECTOR Cover photo: Mussel farm in the Philippines. © FAO/A. Reilly. FAO Fisheries and Aquaculture Circular No. 1165 FIAM/C1165 (En) OVERVIEW OF FOOD FRAUD IN THE FISHERIES SECTOR Alan Reilly Consultant Fisheries and Aquaculture Policy and Resources Division Food and Agriculture Organization FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Rome, 2018 The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations (FAO) concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The mention of specific companies or products of manufacturers, whether or not these have been patented, does not imply that these have been endorsed or recommended by FAO in preference to others of a similar nature that are not mentioned. The views expressed in this information product are those of the author(s) and do not necessarily reflect the views or policies of FAO. ISBN 978-92-5-130402-0 © FAO, 2018 FAO encourages the use, reproduction and dissemination of material in this information product. Except where otherwise indicated, material may be copied, downloaded and printed for private study, research and teaching purposes, or for use in non-commercial products or services, provided that appropriate acknowledgement of FAO as the source and copyright holder is given and that FAO’s endorsement of users’ views, products or services is not implied in any way. -

Fish in Disguise: Seafood Fraud in Korea
Fish in disguise: Seafood fraud in Korea A briefing by the Environmental Justice Foundation 1 Executive summary Between January and December 2018, the Environmental Justice Foundation (EJF) used DNA testing to determine levels of seafood fraud in the Republic of Korea. The results showed that over a third of samples tested were mislabelled. This mislabelling defrauds consumers, risks public health, harms the marine environment and can be associated with serious human rights abuses across the world. These findings demonstrate the urgent need for greater transparency and traceability in Korean seafood, including imported products. Key findings: • Over a third of seafood samples (34.8%, 105 of 302 samples) genetically analysed were mislabelled. • Samples labelled Fleshy Prawn, Fenneropenaeus chinensis (100%), Japanese Eel, Anguilla japonica (67.7%), Mottled Skate, Raja pulchra (53.3%) and Common Octopus, Octopus vulgaris (52.9%) had the highest rates of mislabelling. • Not a single sample labelled Fleshly Prawn was the correct species. • Mislabelling was higher in restaurants, fish markets and online than in general markets or superstores. • By processed types, sushi (53.9%), fresh fish (38.9%) and sashimi (33.6%) were the most likely to be mislabelled. • The seafood fraud identified by this research has direct negative impacts for consumers. It is clear that for some species sampled consumers were likely to be paying more than they should. For example, more than half of the eel and skate samples that were labelled domestic were actually found to be imported, which can cost only half of the price of domestic products. Swordfish mislabelled as Bluefin Tuna can be sold for four to five times as much. -

Eliminating Seafood Fraud: a Fishy Approach to Food Policy
Eliminating Seafood Fraud: A Fishy Approach to Food Policy Coastal Routes Policy Briefs #19-01 Emily De Sousa Department of Geography, Environment, and Geomatics University of Guelph Coastal Routes Policy Briefs Series Editor: Philip A. Loring, PhD www.coastalroutes.org Twitter @Coastal_Routes Coastal Routes is a network of researchers, coastal communities, and non-profit organizations all united by our mission of supporting verdant, sustainable, and just coastal livelihoods and places. We are funded primarily by the Social Sciences and Humanities Research Council of Canada and the Arrell Food Institute at the University of Guelph. Cite As: De Sousa, E. 2019. “Eliminating Seafood Fraud: A Fishy Approach to Food Policy.” Coastal Routes Policy Briefs #19-01. Guelph, ON. © 2019 Emily De Sousa. This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which means that you can use, modify, and distribute this document freely, as long as you attribute the original work to the author and do not use the derivative work for commercial purposes. Summary Seafood mislabeling and fraud is a significant problem for Canada, with impacts that accrue across the supply chain, from producers to consumers. To improve fisheries sustainability, support the Canadian seafood industry, and enable Canadians to achieve a healthful and climate friendly diet, new policy measures are needed to combat seafood fraud, including an update to the Safe Food for Canadians regulations, use of DNA barcoding technology, and improved seafood labeling regulations. Context Seafood is a significant source of protein for nearly 3 billion people around the world and contributes $6 billion to the Canadian economy. -

Monthly Summary of Articles on Food Fraud and Adulteration Retrieved Mainly from the JRC Tool Medisys (
September 2016 Monthly Summary of Articles on Food Fraud and Adulteration Retrieved mainly from the JRC tool Medisys (http://medisys.newsbrief.eu/) Food Fraud Cases 02/09/16 - Bluefin Tuna and Swordfish without labelling discovered in Monflacone, Italy Italy 700 kg of Bluefin Tuna and Swordfish coming supposedly from Sicilia were discovered without any label of origin in Monfalcone in Northern Italy. The products were in bad shape and to be commercialized on Fish the markets of the Friul Venezia Julia region. Mislabelling ANSA 03/09/16 - Eight brands of olive oil failed quality tests in Brazil Brazil Out of 20 brands tested by Proteste (a consumer association), 8 did not meet the quality standards. Four of them contained other vegetable oils (substitution) and another four failed the sensory tests for extra Olive oil virgin olive oil (mislabelling). The organisation requests the withdrawal of the fraudulent products from Substitution the market. Mislabelling Proteste official website Estado de Minas (em.com.br) 10/09/16 - 1176 bottles and 29 000 litres of unlabelled wine seized around Naples Italy The local fraud brigades closed an entire beverage facility following poor hygienic conditions and blatant Wine lack of documentation. Particularly 29 000 litres of bulk wine were lacking any traceability. The value of the whole structure is estimated to 1 million EUR. Mislabelling Origin masking Otto Pagine (Napoli) 16/09/16 - 700 kg of seafood without traceability in Sardinia Italy 700 kg of fish and seafood were seized in a port of Sardinia, lacking any documentation on the origin. Fish The incriminated company, based in Naples, was fined 4500 EUR. -

Download the Report
UNTRACEABLE THE CONSEQUENCES OF CANADA’S POORLY REGULATED SEAFOOD SUPPLY CHAINS UNTRACEABLE OCEANA oceana.cocCANADAeana.ca1 Canada is losing up to $93.8 million in tax revenue each year due to the illicit seafood product trade. EXECUTIVE SUMMARY ................................................................................................ 3 WHAT IS ILLEGAL, UNREPORTED AND UNREGULATED FISHING? ........................................ 5 ECONOMIC COSTS ..................................................................................................... 8 HUMAN COSTS ........................................................................................................ 10 SOCIAL AND ENVIRONMENTAL COSTS ......................................................................... 11 SEAFOOD FRAUD AND MISLABELLING .......................................................................... 12 IMPROVING SEAFOOD TRANSPARENCY ....................................................................... 14 RECOMMENDATIONS ................................................................................................ 16 CONSUMER SUPPORT ............................................................................................... 16 BOAT-TO-PLATE TRACEABILITY : A GOVERMENT COMMITMENT ....................................... 17 REFERENCES ............................................................................................................ 18 Published in November 2020 by Oceana Canada Author: Sayara Thurston DOI: 10.5281/zenodo.4148172 Credit frontUNTRACEABLE -

The Global Reach of Seafood Fraud: a Current Review of the Literature
The Global Reach of Seafood Fraud: a Current Review of the Literature June 2014 Authors: Rachel E. Golden and Kimberly Warner, Ph.D. Seafood fraud, the misrepresentation of seafood, has been discovered all around the world. Seafood fraud can take many forms, including false labeling, species substitution, short-weighting and over-glazing to hide the correct identity, origin or weight of the seafood. Oceana‟s campaign to Stop Seafood Fraud focuses on a particular type of fraud: species substitution. To date, Oceana has conducted six seafood studies, which revealed mislabeling in 20 states and Washington, D.C., as well as in France. Oceana‟s National report, released in 2013, is one of the most widely cited reports about seafood fraud in the media. Notably, many other scientists, governments, students and conservation and consumer organizations have conducted studies focused on seafood fraud, including species substitution. Oceana compiled a review of these studies from around the world to assess the global scope of seafood fraud, mislabeling and species substitutions. In this report, we present major findings from the literature review including general trends and alarming examples about the impact of seafood fraud on our oceans, wallets and health. Major findings from the literature review All studies that have investigated seafood fraud have found it – not a single study has reported 0% fraud overall. The vast majority (91%) of studies focused their sampling at the retail end of the supply chain (restaurants and grocery stores). The few studies that sampled mid-chain were split between landings, distributors, processors, and wholesalers. Along with six Oceana studies, the review covers 67 peer-reviewed studies, seven government reports and 23 news articles for a total of 103 sources. -

Pcr-Based Dgge Identification of Bacteria and Yeasts
ESTABLISHMENT OF A GENETIC DATABASE AND MOLECULAR METHODS FOR THE IDENTIFICATION OF FISH SPECIES AVAILBLE ON THE SOUTH AFRICAN MARKET DONNA-MAREÈ CAWTHORN Dissertation presented for the degree of DOCTOR OF PHILOSOPHY (FOOD SCIENCE) in the Faculty of AgriSciences at Stellenbosch University Promotor: Prof. R.C. Witthuhn Co-promotor: Dr. H.A. Steinman December 2011 Stellenbosch University http://scholar.sun.ac.za ii DECLARATION By submitting this dissertation electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. 08 November 2011 _________________________ ___________________ Donna-Mareè Cawthorn Date Copyright © 2011 Stellenbosch University All rights reserved Stellenbosch University http://scholar.sun.ac.za iii ABSTRACT Consumers have the right to accurate information on the fish products they purchase to enable them to make educated seafood selections that will not endanger their own wellbeing or the wellbeing of the environment. Unfortunately, marine resource scarcity, financial incentives and inadequate or poorly enforced regulations have all promoted the mislabelling of fish species on global markets, the results of which may hold economic, conservation and health consequences. The primary aims of this study were to determine the most commonly available fish species on the South African market, to establish and compare DNA-based methods for the unambiguous identification of these species and to utilise the most applicable methods to evaluate the extent of mislabelling on the local fisheries market. -

Food Research International 121 (2019) 723–729
Food Research International 121 (2019) 723–729 Contents lists available at ScienceDirect Food Research International journal homepage: www.elsevier.com/locate/foodres Survey of mislabelling across finfish supply chain reveals mislabelling both T outside and within Canada ⁎ Hanan R. Shehataa,b, Danielle Bourquea,b, Dirk Steinkeb, Shu Chenc, Robert Hannera,b, a Department of Integrative Biology, University of Guelph, Guelph, ON, Canada b Biodiversity Institute of Ontario, University of Guelph, Guelph, ON N1G 2W1, Canada c Laboratory Services Division, University of Guelph, Guelph, ON, Canada ARTICLE INFO ABSTRACT Keywords: Seafood has become one of the most heavily traded food commodities in the era of globalization. International Seafood seafood supply chains are complex and contend with many difficulties in bringing an enormous variety of Substitution products to market. A major challenge involves accurately labelling products such that they comply with a BOLD diverse set of regulatory frameworks, ranging from country-of-origin through to the final point of consumer sale. DNA barcoding Thanks to DNA barcoding, seafood mislabelling is now recognized as a global problem, with potentially negative Importer impacts on human health, economy and the environment. Mislabelling can result from species misidentification, Retailer Regulatory framework use of inappropriate common names, incomplete and/or out-dated regulatory frameworks, or through market substitution. While prior studies have focused primarily on retail and food service establishments, this study used barcoding to assess rates of finfish mislabelling at multiple points in the supply chain within Ontario, Canada.A total of 203 specimens from 12 key targeted species were collected from varied importers, registered processing plants and retailers in Southern Ontario and identified using DNA barcoding. -

Testing Potential Fish Fraud in Community-Supported Fisheries
University of Vermont ScholarWorks @ UVM UVM Honors College Senior Theses Undergraduate Theses 2016 Testing Potential Fish Fraud in Community-Supported Fisheries Ryan J. Tartre University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/hcoltheses Recommended Citation Tartre, Ryan J., "Testing Potential Fish Fraud in Community-Supported Fisheries" (2016). UVM Honors College Senior Theses. 174. https://scholarworks.uvm.edu/hcoltheses/174 This Honors College Thesis is brought to you for free and open access by the Undergraduate Theses at ScholarWorks @ UVM. It has been accepted for inclusion in UVM Honors College Senior Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. Testing Potential Fish Fraud in Community-Supported Fisheries UVM Honors College Senior Thesis By Ryan Tartre Abstract The seafood industry has long been plagued by the substitution of a species under a false label. Seafood mislabeling is a major concern in the management of fish and marine species. Incorrect labels hamper the ability to estimate stock size effectively, reduce consumer choice, and represent potential health hazards. The rates of seafood fraudulence have been shown to differ across businesses and markets, and in recent years, community-supported fishery programs (CSFs) have sprung up as an alternative to fish markets and grocery stores. Using genetic analysis, I show that 17 out of 41 (41.5%) samples examined from multiple markets in New Hampshire and Maine were fraudulent. The rates of fraudulent labeling differed across species and across markets, with community-supported fishery programs having the lowest levels of fraud (3 out of 10 samples, 30%) followed by restaurants (33%), fish markets (44%), sushi restaurants (50%) and grocery stores (58%). -

Minion Sequencing of Seafood in Singapore Reveals Creatively Labelled Flatfishes, Confused 2 Roe, Pig DNA in Squid Balls, and Phantom Crustaceans
bioRxiv preprint doi: https://doi.org/10.1101/826032; this version posted October 31, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 MinION sequencing of seafood in Singapore reveals creatively labelled flatfishes, confused 2 roe, pig DNA in squid balls, and phantom crustaceans 3 4 Jonathan K. I. Ho1,#, Jayanthi Puniamoorthy1#, Amrita Srivathsan1*, Rudolf Meier1* 5 1. Department of Biological Sciences, National University of Singapore, 14 Science Drive 6 4, Singapore 117543 7 * Corresponding authors, [email protected], [email protected] 8 # First authors 9 Keywords: mislabelling, fraud, food safety, DNA barcoding 10 Abstract 11 Food mislabelling is a growing world-wide problem that is increasingly addressed 12 through the authentication of ingredients via techniques like mass spectrometry or DNA- 13 sequencing. However, traditional DNA sequencing methods are slow, expensive, and 14 require well-equipped laboratories. We here test whether these problems can be 15 overcome through the use of Nanopore sequencing. We sequenced 92 single and 13 16 mixed-species samples bought in supermarkets and restaurants in Singapore which has a 17 large and diverse seafood trade. We successfully obtained DNA barcodes for 94% and 18 100% of the single- and mixed-species products after correcting the numerous 19 sequencing errors of MinION reads with a correction pipeline optimized for DNA 20 barcodes. We find comparatively low levels of clear-cut mislabelling for single-species 21 samples (7.6 %) while the rates are higher for mixed-species samples (38.5 %). -
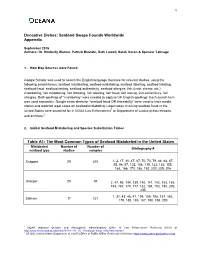
Seafood Swaps Founds Worldwide Appendix Table A1
1 Deceptive Dishes: Seafood Swaps Founds Worldwide Appendix September 2016 Authors: Dr. Kimberly Warner, Patrick Mustain, Beth Lowell, Sarah Geren & Spencer Talmage 1. How Map Sources were Found: Google Scholar was used to search the English language literature for relevant studies, using the following search terms: seafood mislabelling, seafood mislabeling, seafood labelling, seafood labeling, seafood fraud, seafood testing, seafood authenticity, seafood allergies, fish (crab, shrimp, etc.) mislabelling, fish mislabeling, fish labelling, fish labeling, fish fraud, fish testing, fish authenticity, fish allergies. Both spellings of ―mislabeling‖ were needed to capture UK English spellings. Each search term was used separately. Google news alerts for ―seafood fraud OR traceability‖ were used to track media stories and reported legal cases on seafood mislabeling. Legal cases involving seafood fraud in the United States were searched for in NOAA Law Enforcement1 or Department of Justice press releases and archives.2 2. Global Seafood Mislabeling and Species Substitution Tables Table A1: The Most Common Types of Seafood Mislabeled in the United States Mislabeled Number of Number of Bibliography # seafood type studies samples Snapper 29 476 1, 2, 17, 30, 47, 67, 70, 73, 79, 84, 86, 87, 95, 96, 97, 102, 108, 115, 123, 133, 155, 163, 166, 170, 186, 192, 200, 205, 206 Grouper 20 94 2, 47, 95, 134, 139, 140, 141, 142, 153, 155, 163, 167, 174, 177, 182, 184, 192, 194, 200, 205 1, 31, 43, 46, 47, 104, 105, 108, 151, 163, Salmon 17 121 179, 180, 185, 187, 190, 198, 200 1 NOAA (National Oceanic and Atmospheric Administration) Office of Law Enforcement. -
The Seafood Supply Chain from a Fraudulent Perspective
Food Security (2018) 10:939–963 https://doi.org/10.1007/s12571-018-0826-z REVIEW The seafood supply chain from a fraudulent perspective Michaela Fox1 & Mike Mitchell2 & Moira Dean1 & Christopher Elliott 1 & Katrina Campbell1 Received: 27 June 2017 /Accepted: 3 July 2018 /Published online: 8 August 2018 # The Author(s) 2018 Abstract Food fraud is an intentional act for economic gain. It poses a risk to food integrity, the economy, public health and consumers’ ethics. Seafood is one commodity which has endured extensive fraudulent activity owing to its increasing consumer demand, resource limitations, high value and complex supply chains. It is essential that these fraudulent opportunities are revealed, the risk is evaluated and countermeasures for mitigation are assigned. This can be achieved through mapping of the seafood supply chains and identifying the vulnerability analysis critical control points (VACCP), which can be exposed, infiltrated and exploited for fraudulent activity. This research systematically maps the seafood supply chain for three key commodities: finfish, shellfish and crustaceans in the United Kingdom. Each chain is comprised of multiple stakeholders across numerous countries producing a diverse range of products distributed globally. For each supply chain the prospect of fraud, with reference to species substitution, fishery substitution, illegal, unreported and unregulated substitution, species adulteration, chain of custody abuse, catch method fraud, undeclared product extension, modern day slavery and animal welfare, has been identified and evaluated. This mapping of the fraudulent opportunities within the supply chains provides a foundation to rank known and emerging risks and to develop a proactive mitigation plan which assigns control measures and responsibility where vulnerabilities exist.