Studies of Trypanosomiasis in the Luangwa Valley, North-Eastern
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Status, Priorities and Needs for T I Bl Il T I Sustainable Soil Management In
Status, priorities and needs for sustitaina ble so il managemen tit in Zambia SSStalin Sichinga Zamb ia Ag ricu ltu re Resea r ch Institute Introduction Zambia has an area of 750,000 km2 with about 13.9 million people and ample land resources 0ut of 9 million ha cultivable land, only 14% is cropped in any year About 55 - 60% of the land area is covered by natural forest and 6% of Zambia‘s land surface is covered by water. Agro-ecological regions and soil distribution The country is classified into three agro-ecological regions based on soil types, rainfall, and other climatic conditions Agro-Ecological Regions N Chiengi Kaputa Mpulungu W E Nchelenge Mbala Nakonde Mporokoso S Kawambwa Mungwi Isoka Scale 1: 2,500,000 Mwense Luwingu Kasama Chinsali Chilubi Mansa Chama LEGEND Samfya Milenge Mpika Regions Mwinilunga Chililabombwe Solwezi Agro-ecological Region I Chingola Mufulira Lundazi I Ka lul u shi Kitwe Ndola IIa Lufwanyama Luans hya Chavuma Serenje Mambwe Kabompo Masaiti IIb Mpongwe Zambezi Mufumbwe Chipata Kasempa Petauke Katete Chadiza III Annual rainfall is <750mm Kapiri Mposhi Mkushi Nyimba Kabwe Lukulu Kaoma Mumbwa Chibombo Kalabo Mongu Chongwe Lusaka Urban Luangwa Itezhi-Tezhi Kafue Namwala Mazabuka Senanga Monze KEY Siavonga Sesheke Gwembe Shangombo Choma District boundary e Kazungula Kalomo w g n o z a in Livingstone S 200 0 200 400 Kilometers December 2002 The region contains a diversity of soil types ranging from slightly acidic Nitosols to alkaline Luvisols with pockets of Vertisols, Arenosols, Leptosols and, Solonetz. The physical limitations of region I soils Hazards to erosion, lim ite d so il dept h in t he hills an d escarpment zones, presence of hardpans in the pan dambo areas, ppyoor workability in the cracking gy, clay soils, problems of crusting in most parts of the Southern province, low water-holding capacities and the problem of wetness in the valley dambos, plains and swamps. -

Northern Voices - Celebrating 30 Years of Development Partnership in Northern Province, Zambia
Northern Voices - Celebrating 30 years of development partnership in Northern Province, Zambia Mbala Nakonde Isoka Mungwi Luwingu Kasama Chilubi Mpika Lusaka Contents Page Preface 4 Introduction 5 Governance 6 Education 15 Health 23 Water and Sanitation 33 Livelihoods, Food and Nutrition Security 39 HIV & AIDS 49 Preface As Ambassador of Ireland to Zambia, it is with great pleasure that I introduce to you “Northern Voices - Celebrating 30 years of development partnership in Northern Province, Zambia.” This Booklet marks an important milestone in the great friendship I personally had the great pleasure and privilege to work in Northern which has always characterised the relationship between the Province during the years 1996 to 1998, and it is with great pride that I Governments of Ireland and Zambia. 2012 marks the thirtieth return as Ambassador of Ireland to see the page of this great tradition anniversary of the launch of Irish Aid’s local development turning once more, to its next chapter. programme in Zambia’s Northern Province, and presented herewith are thirty distinct perspectives on the nature of that This Booklet offers us the opportunity to reflect on the great many partnership and the many benefits it has engendered – for both successes that we have enjoyed together, while refocusing our energy our great peoples. and determination upon the challenges yet to come. It is my sincere hope that you find it an interesting and valuable resource. The Booklet tells the story of the thirty year programme of development cooperation through the eyes of the very people Finbar O’Brien that have benefitted from it most. -
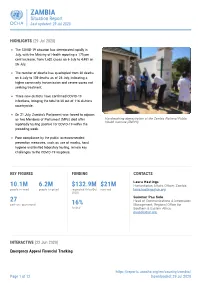
ZAMBIA Situation Report Last Updated: 29 Jul 2020
ZAMBIA Situation Report Last updated: 29 Jul 2020 HIGHLIGHTS (29 Jul 2020) The COVID-19 situation has deteriorated rapidly in July, with the Ministry of Health reporting a 175 per cent increase, from 1,632 cases on 6 July to 4,481 on 26 July. The number of deaths has quadrupled from 30 deaths on 6 July to 139 deaths as of 26 July, indicating a higher community transmission and severe cases not seeking treatment. Three new districts have confirmed COVID-19 infections, bringing the total to 38 out of 116 districts countrywide. On 21 July, Zambia’s Parliament was forced to adjourn as two Members of Parliament (MPs) died after Handwashing demostration at the Zambia National Public Health Institute (ZNPHI) reportedly testing positive for COVID-19 within the preceding week. Poor compliance by the public to recommended prevention measures, such as use of masks, hand hygiene and limited laboratory testing, remain key challenges to the COVID-19 response. KEY FIGURES FUNDING CONTACTS Laura Hastings 10.1M 6.2M $132.9M $21M Humanitarian Affairs Officer, Zambia people in need people targeted requested (May-Oct received [email protected] 2020) Guiomar Pau Sole 27 Head of Communications & Information partners operational 16% Management, Regional Office for funded Southern & Eastern Africa [email protected] INTERACTIVE (22 Jun 2020) Emergency Appeal Financial Tracking https://reports.unocha.org/en/country/zambia/ Page 1 of 12 Downloaded: 29 Jul 2020 ZAMBIA Situation Report Last updated: 29 Jul 2020 View this interactive graphic: https://bit.ly/ZambiaAppeal2020Funding BACKGROUND (29 Jul 2020) Situation Overview Zambia recorded the first case of COVID-19 on 18 March 2020, and as 26 July 4,481 cases had been confirmed with 139 deaths reported, in 38 out of 116 districts countrywide, according to the Ministry of Health (MoH). -

Rp124 Cover.Pmd
LTC Research Paper Land Tenure, Land Markets, and Instituional Transformation in Zambia edited by Michael Roth with the assistance of Steven G. Smith University of Wisconsin-Madison 175 Science Hall 550 North Park Street Madison, WI 53706 http://www.ies.wisc.edu/ltc/ Research Paper LTC Research Paper 124, U.S. ISSN 0084-0815 originally published in October 1995 LAND TENURE, LAND MARKETS, AND INSTITUTIONAL TRANSFORMATION IN ZAMBIA edited by Michael Roth with the assistance of Steven G. Smith All views, interpretations, recommendations, and conclusions expressed in this paper are those of the authors and not necessarily those of the supporting or cooperating organizations. Andy recommendations or suggestions herein doe not represent the official position of the Government of Zambia. LTC Research Paper 124 Prepared for the Land Tenure Center University of Wisconsin-Madison October 1995 Copyright ® 1995 by the authors. All rights reserved. Readers may make verbatim copies of this document for non-commercial purposes by any means, provided that this copyright notice appears on all such copies. TABLE OF CONTENTS Page Lists of Tables and Figures vii List of Acronyms x Preface xi Chapter 1: Legal Framework and Administration of Land Policy in Zambia 1 I. Introduction 1 A. Issues 1 B. Outline of report 2 II. Colonial policy and settlement 4 III. Agrarian structure 5 IV. Arable land and crop expansion 7 V. Land tenure 14 A. Tenure on State Lands 15 B. Reserve and Trust Land administration 18 C. Urban, housing, and improvement areas 22 D. Agencies responsible for land policy 23 E. Subdivisions 24 VI. -
C:\Users\Public\Documents\GP JOBS\Gazette No. 73 of Friday, 16Th
REPUBLIC OF ZAMBIA Price: K5 net Annual Subscription: Within Lusaka—K200.00 Published by Authority Outside Lusaka—K230.00 No. 6430] Lusaka, Friday, 16th October, 2015 [Vol. LI, No. 73 GAZETTE NOTICE NO. 643 OF 2015 [5929855/13 Zambia Information and Communications Technologies Authority The Information and Communications Technologies Act, 2009 (Act No. 15 of 2009) Notice of Determination of Unserved and Underserved Areas Section 70 (2) of the Information and Communication TechnologiesAct No. 15 of 2009 (ICTAct) empowers the Zambia Information and Communications Technology Authority (ZICTA) to determine a system to promote the widespread availability and usage of electronic communications networks and services throughout Zambia by encouraging the installation of electronic communications networks and the provision for electronic communications services in unserved and underserved areas and communities. Further, Regulation 5 (2) of Statutory Instrument No. 38 of 2012 the Information and Communications Technologies (Universal Access) Regulations 2012 mandates the Authority to designate areas as universal service areas by notice in the gazette. In accordance with the said regulations, the Authority hereby notifies members of the public that areas contained in the Schedule Hereto are hereby designated as universal service areas. M. K. C. MUDENDA (MRS.) Director General SN Site Name Longtitude Latitude Elevation Province 1 Nalusanga_Chunga Headquarter Offices 27.22415 -15.22135 1162 Central 2 Mpusu_KankamoHill 27.03507 -14.45675 1206 Central -

Observing the 2001 Zambia Elections
SPECIAL REPORT SERIES THE CARTER CENTER WAGING PEACE ◆ FIGHTING DISEASE ◆ BUILDING HOPE OBSERVING THE 2001 ZAMBIA ELECTIONS THE CARTER CENTER STRIVES TO RELIEVE SUFFERING BY ADVANCING PEACE AND HEALTH WORLDWIDE; IT SEEKS TO PREVENT AND RESOLVE CONFLICTS, ENHANCE FREEDOM AND DEMOCRACY, AND PROTECT AND PROMOTE HUMAN RIGHTS WORLDWIDE. THE CARTER CENTER NDINDI OBSERVING THE 2001 ZAMBIA ELECTIONS OBSERVING THE 2001 ZAMBIA ELECTIONS FINAL REPORT THE CARTER CENTER The Democracy Program One Copenhill Atlanta, GA 30307 (404) 420-5188 FAX (404) 420-5196 WWW.CARTERCENTER.ORG OCTOBER 2002 1 THE CARTER CENTER NDI OBSERVING THE 2001 ZAMBIA ELECTIONS 2 THE CARTER CENTER NDINDI OBSERVING THE 2001 ZAMBIA ELECTIONS TABLE OF CONTENTS Carter Center Election Observation Delegation and Staff ............................................................... 5 Terms and Abbreviations ................................................................................................................. 7 Foreword ......................................................................................................................................... 8 Executive Summary .......................................................................................................................... 10 Acknowledgments............................................................................................................................. 15 Background ...................................................................................................................................... -

Rainy Season Forecast 2019/2020
REPUBLIC OF ZAMBIA Ministry of Transport and Communications ZAMBIA METEOROLOGICAL DEPARTMENT RAINY SEASON FORECAST 2019/2020 Issued by Edson NKONDE Director of Meteorology Telephone: +260 211 251912 TeleFax: +260 211 252728 P.O. Box 30200, Lusaka E-mail: [email protected] Web Site: www.zmd.gov.zm 4th September, 2019 GENERAL SITUATION Generally, much of Zambia is likely to receive Normal to Above Normal rainfall during the 2019/2020 rainfall season. However, there is a likelihood of dry spells around February especially over the Southern half of the country. DETAILED FORECAST October, November and December (OND): There is a high chance of receiving Normal to Above Normal rainfall for most parts of Zambia except for the north-eastern areas (Isoka, Chama, Mpika, Mbala, Nakonde, Mpulungu, Chinsali, Mungwi and Lundazi districts) which are likely to receive Normal to Below Normal rainfall (Figure 1). November, December and January (NDJ): The southern half of the country is likely to receive Above Normal Rainfall while the northern half has a high chance of receiving Normal to Above Normal rainfall with the exception of the north-eastern areas where there is a likelihood of Normal to Below Normal rainfall (Figure 2). December, January and February (DJF): There is a high likelihood of receiving Normal to Above Normal rainfall for the southern half of the country while the northern half has a high chance of recording Above Normal Rainfall (Figure 3). January, February and March (JFM): The northern half of the country has a high chance of receiving Normal to Above Normal Rainfall while the southern half is likely to received Normal to Below Normal Rainfall. -

Consultancy Services for Techno Studies, Detailed Engineering
Consultancy Services for Techno Studies, Detailed Engineering Design and Preparation of THE REPUBLIC Tender Documents for the Rehabilitation of the OF ZAMBIA AFRICAN Chinsali-Nakonde Road (T2) DEVELOPMENT Draft ESIA Report BANK Consultancy Services for Techno Studies, Detailed Engineering Design and Preparation of Tender Documents for the Rehabilitation of the Chinsali-Nakonde Road (T2) Draft Environmental Impact Statement (EIS) Issue No: 01 First Date of Issue: January 2015 Second Date of Issue: February 2015 Prepared by: Chamfya Moses Quality Control: Chansa Davies Approved: Mushinge Renatus i Consultancy Services for Techno Studies, Detailed Engineering Design and Preparation of THE REPUBLIC Tender Documents for the Rehabilitation of the OF ZAMBIA AFRICAN Chinsali-Nakonde Road (T2) DEVELOPMENT Draft ESIA Report BANK DECLARATION: DEVELOPER I, , on behalf of the Road Development Agency of Zambia, hereby submit this Draft Environmental and Social Impact Statement for the proposed rehabilitation of the 210Km T2 Road from Chinsali to Nakonde Roads in accordance with the Environmental Management Act 2011 and the Environmental Impact Assessment Regulations S.I. No. 28 of 1997. Signed at LUSAKA on this day of , 2015 Signature: Designation: ROAD DEVELOPMENT AGENCY DECLARATION: CONSULTING ENGINEER I, , on behalf of BICON (Z) Ltd, hereby submit this Draft Environmental and Social Impact Statement for the proposed rehabilitation of the 210Km T2 Road from Chinsali to Nakonde Road in accordance with the Environmental Management Act 2011 and the -

Livelihood Zones Analysis Zambia
Improved livelihoods for smallholder farmers LIVELIHOOD ZONES ANALYSIS A tool for planning agricultural water management investments Zambia Prepared by Mukelabai Ndiyoi & Mwase Phiri, Farming Systems Association of Zambia (FASAZ), Lusaka, Zambia, in consultation with FAO, 2010 About this report The AgWater Solutions Project aimed at designing agricultural water management (AWM) strategies for smallholder farmers in sub Saharan Africa and in India. The project was managed by the International Water Management Institute (IWMI) and operated jointly with the Food and Agriculture Organization of the United Nations (FAO), International Food Policy Research Institute (IFPRI), the Stockholm Environmental Institute (SEI) and International Development Enterprise (IDE). It was implemented in Burkina Faso, Ethiopia, Ghana, Tanzania, Zambia and in the States of Madhya Pradesh and West Bengal in India. Several studies have highlighted the potential of AWM for poverty alleviation. In practice, however, adoption rates of AWM solutions remain low, and where adoption has taken place locally, programmes aimed at disseminating these solutions often remain a challenge. The overall goal of the project was to stimulate and support successful pro-poor, gender-equitable AWM investments, policies and implementation strategies through concrete, evidence-based knowledge and decision-making tools. The project has examined AWM interventions at the farm, community, watershed, and national levels. It has analyzed opportunities and constraints of a number of small-scale AWM interventions in several pilot research sites across the different project countries, and assessed their potential in different agro-climatic, socio-economic and political contexts. This report was prepared as part of the efforts to assess the potential for AWM solutions at national level. -

Registered Voters by Gender and Constituency
REGISTERED VOTERS BY GENDER AND CONSTITUENCY % OF % OF SUB % OF PROVINCIAL CONSTITUENCY NAME MALES MALES FEMALES FEMALES TOTAL TOTAL KATUBA 25,040 46.6% 28,746 53.4% 53,786 8.1% KEEMBE 23,580 48.1% 25,453 51.9% 49,033 7.4% CHISAMBA 19,289 47.5% 21,343 52.5% 40,632 6.1% CHITAMBO 11,720 44.1% 14,879 55.9% 26,599 4.0% ITEZH-ITEZHI 18,713 47.2% 20,928 52.8% 39,641 5.9% BWACHA 24,749 48.1% 26,707 51.9% 51,456 7.7% KABWE CENTRAL 31,504 47.4% 34,993 52.6% 66,497 10.0% KAPIRI MPOSHI 41,947 46.7% 47,905 53.3% 89,852 13.5% MKUSHI SOUTH 10,797 47.3% 12,017 52.7% 22,814 3.4% MKUSHI NORTH 26,983 49.5% 27,504 50.5% 54,487 8.2% MUMBWA 23,494 47.9% 25,545 52.1% 49,039 7.4% NANGOMA 12,487 47.4% 13,864 52.6% 26,351 4.0% LUFUBU 5,491 48.1% 5,920 51.9% 11,411 1.7% MUCHINGA 10,072 49.7% 10,200 50.3% 20,272 3.0% SERENJE 14,415 48.5% 15,313 51.5% 29,728 4.5% MWEMBEZHI 16,756 47.9% 18,246 52.1% 35,002 5.3% 317,037 47.6% 349,563 52.4% 666,600 100.0% % OF % OF SUB % OF PROVINCIAL CONSTITUENCY NAME MALES MALES FEMALES FEMALES TOTAL TOTAL CHILILABOMBWE 28,058 51.1% 26,835 48.9% 54,893 5.4% CHINGOLA 34,695 49.7% 35,098 50.3% 69,793 6.8% NCHANGA 23,622 50.0% 23,654 50.0% 47,276 4.6% KALULUSHI 32,683 50.1% 32,614 49.9% 65,297 6.4% CHIMWEMWE 29,370 48.7% 30,953 51.3% 60,323 5.9% KAMFINSA 24,282 51.1% 23,214 48.9% 47,496 4.6% KWACHA 31,637 49.3% 32,508 50.7% 64,145 6.3% NKANA 27,595 51.9% 25,562 48.1% 53,157 5.2% WUSAKILE 23,206 50.5% 22,787 49.5% 45,993 4.5% LUANSHYA 26,658 49.5% 27,225 50.5% 53,883 5.3% ROAN 15,921 50.1% 15,880 49.9% 31,801 3.1% LUFWANYAMA 18,023 50.2% -

Members of the Northern Rhodesia Legislative Council and National Assembly of Zambia, 1924-2021
NATIONAL ASSEMBLY OF ZAMBIA Parliament Buildings P.O Box 31299 Lusaka www.parliament.gov.zm MEMBERS OF THE NORTHERN RHODESIA LEGISLATIVE COUNCIL AND NATIONAL ASSEMBLY OF ZAMBIA, 1924-2021 FIRST EDITION, 2021 TABLE OF CONTENTS FOREWORD ................................................................................................................................................ 3 PREFACE ..................................................................................................................................................... 4 ACKNOWLEDGEMENTS .......................................................................................................................... 5 ABBREVIATIONS ...................................................................................................................................... 7 INTRODUCTION ........................................................................................................................................ 9 PART A: MEMBERS OF THE LEGISLATIVE COUNCIL, 1924 - 1964 ............................................... 10 PRIME MINISTERS OF THE FEDERATION OF RHODESIA .......................................................... 12 GOVERNORS OF NORTHERN RHODESIA AND PRESIDING OFFICERS OF THE LEGISTRATIVE COUNCIL (LEGICO) ............................................................................................... 13 SPEAKERS OF THE LEGISTRATIVE COUNCIL (LEGICO) - 1948 TO 1964 ................................. 16 DEPUTY SPEAKERS OF THE LEGICO 1948 TO 1964 .................................................................... -

Chiefdoms/Chiefs in Zambia
CHIEFDOMS/CHIEFS IN ZAMBIA 1. CENTRAL PROVINCE A. Chibombo District Tribe 1 HRH Chief Chitanda Lenje People 2 HRH Chieftainess Mungule Lenje People 3 HRH Chief Liteta Lenje People B. Chisamba District 1 HRH Chief Chamuka Lenje People C. Kapiri Mposhi District 1 HRH Senior Chief Chipepo Lenje People 2 HRH Chief Mukonchi Swaka People 3 HRH Chief Nkole Swaka People D. Ngabwe District 1 HRH Chief Ngabwe Lima/Lenje People 2 HRH Chief Mukubwe Lima/Lenje People E. Mkushi District 1 HRHChief Chitina Swaka People 2 HRH Chief Shaibila Lala People 3 HRH Chief Mulungwe Lala People F. Luano District 1 HRH Senior Chief Mboroma Lala People 2 HRH Chief Chembe Lala People 3 HRH Chief Chikupili Swaka People 4 HRH Chief Kanyesha Lala People 5 HRHChief Kaundula Lala People 6 HRH Chief Mboshya Lala People G. Mumbwa District 1 HRH Chief Chibuluma Kaonde/Ila People 2 HRH Chieftainess Kabulwebulwe Nkoya People 3 HRH Chief Kaindu Kaonde People 4 HRH Chief Moono Ila People 5 HRH Chief Mulendema Ila People 6 HRH Chief Mumba Kaonde People H. Serenje District 1 HRH Senior Chief Muchinda Lala People 2 HRH Chief Kabamba Lala People 3 HRh Chief Chisomo Lala People 4 HRH Chief Mailo Lala People 5 HRH Chieftainess Serenje Lala People 6 HRH Chief Chibale Lala People I. Chitambo District 1 HRH Chief Chitambo Lala People 2 HRH Chief Muchinka Lala People J. Itezhi Tezhi District 1 HRH Chieftainess Muwezwa Ila People 2 HRH Chief Chilyabufu Ila People 3 HRH Chief Musungwa Ila People 4 HRH Chief Shezongo Ila People 5 HRH Chief Shimbizhi Ila People 6 HRH Chief Kaingu Ila People K.