(3Dxms) Chemical Images Interactive Spatio-Spectral
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Near-Infrared Chemical Imaging and the PAT Initiative NIR-CI Adds a Completely New Dimension to Conventional NIR Spectroscopy
Molecular Spectroscopy Workbench Near-infrared Chemical Imaging and the PAT Initiative NIR-CI adds a completely new dimension to conventional NIR spectroscopy. E. Neil Lewis, Joe Schoppelrei, and Eunah Lee Dear and gentle readers, this month I present an important other manufacturing steps have on the final dosage form? To tool in the process analytical technologies (PAT) initiative: encourage the PAT initiative FDA is streamlining the mecha- Near-infrared chemical imaging (NIR-CI). You might wonder nism for adopting new technologies in pharmaceutical manu- why I am seemingly concentrating on IR and NIR. The an- facturing. swer is quite simple: I need all the heat I can get, here in New York. Seriously, E. Neil Lewis, Joe Schoppelrei, and The Role of Near-infrared Chemical Imaging (NIR-CI) Eunah Lee of Spectral Dimensions (Olney, MD) have put to- A typical tablet is not just a pressed block of a single material, gether an excellent explanation of what NIR-CI is and what but rather a complex matrix containing one or more active its part in PAT is and will be. For those of you who do not pharmaceutical ingredients (APIs), fillers, binders, disinte- know Neil Lewis, he is one of the pioneers in IR and NIR im- grants, lubricants, and other materials. A basic problem in aging. He has won numerous awards for his research at the pharmaceutical manufacturing is that a relatively simple for- National Institutes of Health (NIH) and subsequent accom- mulation with identical ingredients can produce widely vary- plishments. It is truly exciting to have this paper in my hum- ing therapeutic performance depending upon how the ingre- ble column and I know you will find it informative and enter- dients are distributed in the final matrix. -

Comprehensive Bibliography on Martian Meteorites (Compiled by C
Comprehensive Bibliography on Martian Meteorites (compiled by C. Meyer, March 2008) Abu Aghreb A.E., Ghadi A.M., Schlüter J., Schultz L. and Thiedig F. (2003) Hamadah al Hamra and Dar al Gani: A comparison of two meteorite fields in the Libyan Sahara (abs). Meteoritics & Planet. Sci. 38, A48. Agee Carl B. (2002) Garnet and majorite fractionation in the early Earth and Mars (abs#1862). Lunar Planet. Sci. XXXIII Lunar Planetary Institute, Houston. (CD-ROM). (see address of LPI in Appendix III) Agee C.B., Bogard Don D., Draper D.S., Jones J.H., Meyer Chuck and Mittlefehldt D.W. (2000) Proposed science requirements and acquisition priorities for the first Mars sample return (abs). In Concept and Approaches for Mars Exploration. Part 1 (ed. S. Hubbard) LPI Contribution # 1062. Lunar Planetary Institute, Houston. Agee C.B. and Draper Dave S. (2003) Melting of model Martian mantle at high pressure: Implications for the composition of the Martian basalt source region (abs#1408). Lunar Planet. Sci. Conf. 34th, Lunar Planetary Institute, Houston (CD-ROM). Agerkvist D.P. and Vistisen L. (1993) Mössbauer spectroscopy of the SNC meteorite Zagami (abs). Lunar Planet. Sci. XXIV, 1-2. Lunar Planetary Institute, Houston. Zagami Agerkvist D.P., Vistisen L., Madsen M.B. and Knudsen J.M. (1994) Magnetic properties of Zagami and Nakhla (abs). Lunar Planet. Sci. XXV, 1-2. Lunar Planetary Institute, Houston. Zagami Nakhla Akai J. (1997) Characteristics of iron-oxide and iron-sulfide grains in meteorites and terrestrial sediments, with special references to magnetite grains in Allan Hills 84001 (abs). Meteoritics & Planet. Sci. -

Army Acquisition Workforce Dependency on E-Mail for Formal
ARMY ACQUISITION WORKFORCE DEPENDENCY ON E-MAIL FOR FORMAL WORK COORDINATION: FINDINGS AND OPPORTUNITIES FOR WORKFORCE PERFORMANCE IMPROVEMENT THROUGH E-MAIL-BASED SOCIAL NETWORK ANALYSIS KENNETH A. LORENTZEN May 2013 PUBLISHED BY THE DEFENSE ACQUISITION UNIVERSITY PRESS PROJECT ADVISOR: BOB SKERTIC CAPITAL AND NORTHEAST REGION, DAU THE SENIOR SERVICE COLLEGE FELLOWSHIP PROGRAM ABERDEEN PROVING GROUND, MD PAGE LEFT BLANK INTENTIONALLY .ARMY ACQUISITION WORKFORCE DEPENDENCY ON E-MAIL FOR FORMAL WORK COORDINATION: FINDINGS AND OPPORTUNITIES FOR WORKFORCE PERFORMANCE IMPROVEMENT THROUGH E-MAIL-BASED SOCIAL NETWORK ANALYSIS KENNETH A. LORENTZEN May 2013 PUBLISHED BY THE DEFENSE ACQUISITION UNIVERSITY PRESS PROJECT ADVISOR: BOB SKERTIC CAPITAL AND NORTHEAST REGION, DAU THE SENIOR SERVICE COLLEGE FELLOWSHIP PROGRAM ABERDEEN PROVING GROUND, MD PAGE LEFT BLANK INTENTIONALLY ii Table of Contents Table of Contents ............................................................................................................................ ii List of Figures ................................................................................................................................ vi Abstract ......................................................................................................................................... vii Chapter 1—Introduction ................................................................................................................. 1 Background and Motivation ................................................................................................. -

Inviwo — a Visualization System with Usage Abstraction Levels
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS, VOL X, NO. Y, MAY 2019 1 Inviwo — A Visualization System with Usage Abstraction Levels Daniel Jonsson,¨ Peter Steneteg, Erik Sunden,´ Rickard Englund, Sathish Kottravel, Martin Falk, Member, IEEE, Anders Ynnerman, Ingrid Hotz, and Timo Ropinski Member, IEEE, Abstract—The complexity of today’s visualization applications demands specific visualization systems tailored for the development of these applications. Frequently, such systems utilize levels of abstraction to improve the application development process, for instance by providing a data flow network editor. Unfortunately, these abstractions result in several issues, which need to be circumvented through an abstraction-centered system design. Often, a high level of abstraction hides low level details, which makes it difficult to directly access the underlying computing platform, which would be important to achieve an optimal performance. Therefore, we propose a layer structure developed for modern and sustainable visualization systems allowing developers to interact with all contained abstraction levels. We refer to this interaction capabilities as usage abstraction levels, since we target application developers with various levels of experience. We formulate the requirements for such a system, derive the desired architecture, and present how the concepts have been exemplary realized within the Inviwo visualization system. Furthermore, we address several specific challenges that arise during the realization of such a layered architecture, such as communication between different computing platforms, performance centered encapsulation, as well as layer-independent development by supporting cross layer documentation and debugging capabilities. Index Terms—Visualization systems, data visualization, visual analytics, data analysis, computer graphics, image processing. F 1 INTRODUCTION The field of visualization is maturing, and a shift can be employing different layers of abstraction. -

Image Processing on Optimal Volume Sampling Lattices
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1314 Image processing on optimal volume sampling lattices Thinking outside the box ELISABETH SCHOLD LINNÉR ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 ISBN 978-91-554-9406-3 UPPSALA urn:nbn:se:uu:diva-265340 2015 Dissertation presented at Uppsala University to be publicly examined in Pol2447, Informationsteknologiskt centrum (ITC), Lägerhyddsvägen 2, hus 2, Uppsala, Friday, 18 December 2015 at 10:00 for the degree of Doctor of Philosophy. The examination will be conducted in English. Faculty examiner: Professor Alexandre Falcão (Institute of Computing, University of Campinas, Brazil). Abstract Schold Linnér, E. 2015. Image processing on optimal volume sampling lattices. Thinking outside the box. (Bildbehandling på optimala samplingsgitter. Att tänka utanför ramen). Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1314. 98 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-554-9406-3. This thesis summarizes a series of studies of how image quality is affected by the choice of sampling pattern in 3D. Our comparison includes the Cartesian cubic (CC) lattice, the body- centered cubic (BCC) lattice, and the face-centered cubic (FCC) lattice. Our studies of the lattice Brillouin zones of lattices of equal density show that, while the CC lattice is suitable for functions with elongated spectra, the FCC lattice offers the least variation in resolution with respect to direction. The BCC lattice, however, offers the highest global cutoff frequency. The difference in behavior between the BCC and FCC lattices is negligible for a natural spectrum. We also present a study of pre-aliasing errors on anisotropic versions of the CC, BCC, and FCC sampling lattices, revealing that the optimal choice of sampling lattice is highly dependent on lattice orientation and anisotropy. -

IUS 2020 Table of Contents
IUS 2020 Table of Contents IUS 2020 Table of Contents .................................................................................................................................1 IUS 2020 Patrons & Sponsor ..............................................................................................................................2 IUS 2020 Short Courses .......................................................................................................................................5 IUS 2020 Opening/Closing Events ....................................................................................................................7 IUS 2020 Live Social/Workshop Events ...........................................................................................................7 Technical Program Table of Contents .......................................................................................................... 14 1 IUS 2020 Patrons & Sponsor Platinum Patron Verasonics designs and markets leading-edge Vantage™ ultrasound research systems for academic and commercial investigators. These real-time, software-based, programmable ultrasound systems accelerate research by providing unsurpassed speed and control to simplify the data collection and analysis process. Researchers in 34 countries routinely use the unparalleled flexibility of the Vantage platform to advance the art and science of ultrasound through their own research efforts. In addition, to protect your investment and encompass additional research options, every Vantage -

Medline-Sourced Journals Are Indicated in Green) Print-ISSN E-ISSN
Sourcerecord id Source Title (Medline-sourced journals are indicated in Green) Print-ISSN E-ISSN 18500162600 21st Century Music 15343219 21100404576 2D Materials 20531583 21100447128 3 Biotech 2190572X 21905738 21100779062 3D Printing and Additive Manufacturing 23297662 23297670 21100229836 3D Research 20926731 19700200922 3L: Language, Linguistics, Literature 01285157 145295 4OR 16194500 16142411 16400154734 A + U-Architecture and Urbanism 03899160 5700161051 A Contrario 16607880 21100399164 A&A case reports 23257237 21100881366 A&A practice 25753126 19600162043 A.M.A. American Journal of Diseases of Children 00968994 19400157806 A.M.A. archives of dermatology 00965359 19600162081 A.M.A. Archives of Dermatology and Syphilology 00965979 19400157807 A.M.A. archives of industrial health 05673933 19600162082 A.M.A. Archives of Industrial Hygiene and Occupational Medicine 00966703 19400157808 A.M.A. archives of internal medicine 08882479 19400158171 A.M.A. archives of neurology 03758540 19400157809 A.M.A. archives of neurology and psychiatry 00966886 19400157810 A.M.A. archives of ophthalmology 00966339 19400157811 A.M.A. archives of otolaryngology 00966894 19400157812 A.M.A. archives of pathology 00966711 19400157813 A.M.A. archives of surgery 00966908 21100456161 a/b: Auto/Biography Studies 21517290 11600153683 A|Z ITU Journal of Faculty of Architecture 13028324 21100780699 A+BE Architecture and the Built Environment 22123202 22147233 5800207606 AAA, Arbeiten aus Anglistik und Amerikanistik 01715410 28033 AAC: Augmentative and Alternative -
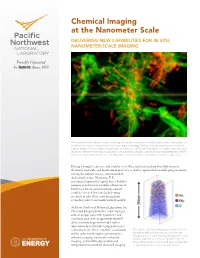
Chemical Imaging at the Nanometer Scale Delivering New Capabilities for in Situ, Nanometer-Scale Imaging
Chemical Imaging at the Nanometer Scale DELIVERING NEW CAPABILITIES FOR IN SITU, NANOMETER-ScALE IMAGING The majority of microbes in natural and engineered environments live within structured communities or biofilms (seen here using confocal laser scanning microscopy). Biofilms include a poorly characterized organic matrix, termed extracellular polymeric substance (EPS), which facilitates certain biogeochemical reactions. We are enhancing visualization, compositional analysis, and functional characterization of EPS to better understand its influence on subsurface reactions vital to environmental and energy issues. Having a complete, precise, and realistic view of the molecular interactions that occur in chemical, materials, and biochemical processes is vital to exponential scientific progress and to solving the nation’s energy, environmental, and security issues. Numerous U.S. government-sponsored reports have identified imaging as critical for scientific advancement. However, current instrumentation cannot reach the needed level of clarity, leaving scientists to infer what is occurring from secondary sources and mathematical models. At Pacific Northwest National Laboratory, the Chemical Imaging Initiative is developing a suite of unique tools with nanometer-scale resolution and element specificity that will allow scientists to go from model system observation to real-world manipulation on a molecular level. These real-time, in situ tools Through the Chemical Imaging Initiative at Pacific will be achieved through near-nanometer Northwest -

Low Wnt/B-Catenin Signaling Determines Leaky Vessels in The
RESEARCH ARTICLE Low wnt/b-catenin signaling determines leaky vessels in the subfornical organ and affects water homeostasis in mice Fabienne Benz1, Viraya Wichitnaowarat1, Martin Lehmann1, Raoul FV Germano2, Diana Mihova1, Jadranka Macas1, Ralf H Adams3, M Mark Taketo4, Karl-Heinz Plate1,5,6,7,8, Sylvaine Gue´ rit1, Benoit Vanhollebeke2,9, Stefan Liebner1,5,6* 1Institute of Neurology (Edinger Institute), University Hospital, Goethe University Frankfurt, Frankfurt am Main, Germany; 2Laboratory of Neurovascular Signaling, Department of Molecular Biology, ULB Neuroscience Institute, Universite´ libre de Bruxelles, Bruxelles, Belgium; 3Department of Tissue Morphogenesis, Max-Planck- Institute for Molecular Biomedicine, University of Mu¨ nster, Faculty of Medicine, Mu¨ nster, Germany; 4Division of Experimental Therapeutics, Graduate School of Medicine, Kyoto University, Kyoto, Japan; 5Excellence Cluster Cardio-Pulmonary systems (ECCPS), Partner site Frankfurt, Frankfurt, Germany; 6German Cancer Consortium (DKTK), Partner Site Frankfurt/Mainz, Frankfurt, Germany; 7German Center for Cardiovascular Research (DZHK), Partner site Frankfurt/Mainz, Frankfurt, Germany; 8German Cancer Research Center (DKFZ), Heidelberg, Germany; 9Walloon Excellence in Life Sciences and Biotechnology (WELBIO), Wallonia, Belgium Abstract The circumventricular organs (CVOs) in the central nervous system (CNS) lack a vascular blood-brain barrier (BBB), creating communication sites for sensory or secretory neurons, involved in body homeostasis. Wnt/b-catenin signaling is essential for BBB development and *For correspondence: [email protected] maintenance in endothelial cells (ECs) in most CNS vessels. Here we show that in mouse development, as well as in adult mouse and zebrafish, CVO ECs rendered Wnt-reporter negative, Competing interests: The suggesting low level pathway activity. Characterization of the subfornical organ (SFO) vasculature authors declare that no revealed heterogenous claudin-5 (Cldn5) and Plvap/Meca32 expression indicative for tight and competing interests exist. -

Syxcnnorron Raor.Tnox Iwrnannp Spbcrnomrcroscopy: A
SyxcnnorRoN Raor.tnox Iwrnannp SpBcrnoMrcRoscoPY:A Noxnrvesrvp CnBurcAL Pnosp FoR MomroRrNG BrocnocHEMrcar PnocESSES H.-Y. N. Holmanl'2 and M. C. Martins rEcology Department, Earth SciencesDivision, Lawrence Berkeley National Laboratory Universi6 of California, Berkelev. Calif ornta 947 2 0 2Vrtual Institute foi Microbial Stressand Sun-ival, Lawrence Berkeley National Laboratory lJniversin' of California, Berkelev. Calif ornta 9 47 2 0 rAduanced Light Sou... Division. Lav-rence Berkeley National Laboratory Universiw of California, Berkelev. California 947 20 L lntroduction IL SR-FTIR Spectromicroscopl, A. Background B. SynchrotronIR Light Sources C. SynchrotronIR Spectromicroscopyof BiogeochemicalSystems III. BiogeochemicalProcesses Measured by SR-FTIR Spectromicroscopy A. Instrumentation B. SpectralAnalysis C. Application Examples I\'. Future Possibilitiesand Requirements Acknowledgments References A long-standingdesire in biogeochemistryis to be abie to examinethe clcling of elementsby microorganisms.as the processesare happening on surfacesof earth and environmentalmaterials. Over the past decade, physics,engineering, and instrumentationinnovations have led to the intro- duction of srnchrotron radiation-basedinfrared (IR) spectromicroscopy. Spatial resolutionsof lessthan 10 micrometers(pm) and photon energies of lessthan an electronvolt make s\nchrotron IR spectromicroscop)non- invasive and useful for follo$-in_slhe course of biogeochemicalprocesses on complex heterogeneoussurfaces of earth and environmentalmaterials. In this chapter,we will first briefll describethe technologyand then present ;9 AtLan;t-' tn.1'qranmy, Volume 90 Copgight 1006. F.lse\ierInc. All rights resenerl. 006i-I ltl06 $15.00 DOI: I 0. I 0 I 6/50065-2I 1l(06)90003 -0 80 H.-Y. N. HOLMAN AND M. C. MARTIN several examples demonstrating its application potentials in probing and imaging biogeochemicalprocesses. Q 2006,Elsevier Inc. L INTRODUCTION Microorganisms are important agents in the geochemicalcycling of ele- ments. -

Atmosphere Bibliography
™ POSEIDON SELECT Pchips BIBLIOGRAPHY — JOURNAL ARTICLES Quantifiably Better™ Title Authors Citations Web Link Aliyah, Kinanti;Lyu, Jieli;Goldmann, Claire;Bizien, Thomas;Hamon, Real-Time In Situ Observations Reveal Aliyah, Kinanti;Lyu, Jieli;Goldmann, Claire;Bizien, Cyrille;Alloyeau, Damien;Constantin, Doru , Real-Time In Situ Observations a Double Role for Ascorbic Acid in the Thomas;Hamon, Cyrille;Alloyeau, Damien;Constantin, Reveal a Double Role for Ascorbic Acid in the Anisotropic Growth of Silver https://doi.org/10.1021/acs.jpclett.0c00121 Anisotropic Growth of Silver on Gold Doru on Gold, 2020, The Journal of Physical Chemistry Letters, 10.1021/acs. jpclett.0c00121 Kazemi, Mohammad Ali Akhavan;Raval, Parth;Cherednichekno, Molecular-Level Insight into Correlation Kazemi, Mohammad Ali Akhavan;Raval, Kirill;Chotard, Jean-Noel;Krishna, Anurag;Demortiere, Arnaud;Reddy, G. N. between Surface Defects and Stability Parth;Cherednichekno, Kirill;Chotard, Jean- Manjunatha;Sauvage, Frédéric , Molecular-Level Insight into Correlation https://onlinelibrary.wiley.com/doi/abs/10.1002/ of Methylammonium Lead Halide Noel;Krishna, Anurag;Demortiere, Arnaud;Reddy, G. N. between Surface Defects and Stability of Methylammonium Lead Halide smtd.202000834 Perovskite Under Controlled Humidity Manjunatha;Sauvage, Frédéric Perovskite Under Controlled Humidity, 2020, Small Methods, https://doi. org/10.1002/smtd.202000834 Li, Weiya;Liu, Bin;Hubert, Céline;Perro, Adeline;Duguet, Etienne;Ravaine, Self-assembly of colloidal polymers Li, Weiya;Liu, Bin;Hubert, Céline;Perro, -

Chemical Imaging
Chemical Imaging Chemical Imaging (CI) combines different technologies like optical microscopy, digital imaging and molecular spectroscopy in combination with multivariate data analy- sis methods. CI systems can be classified in Whiskbroom- (Mapping), Staring- (2D wavelength scanning) and Pushbroom- (line scanning with spectral dispersion) im- Whiskbroom agers. The concept can be applied over a wide spectro- scopic range from UV-VIS-NIR, fluorescence, IR and Ra- man. Whiskbroom Imaging Whiskbroom Imaging means that each single point of the sample is spectrally recorded with a single detector. Such systems have a broad flexibility in terms of sam- ple size, raster width, spectral ranges and implementa- - high spectral resolution tion in optical methods. This mode leads to a band- - high discrimination power interleaved-pixel (BIP) data format. Whiskbroom Imag- - high flexibility ing is typical for IR, Raman and confocal microscopes. Staring Staring Imaging Staring Imaging means that a sequence of two- dimensional images of a fixed sample at different wave- lengths is acquired. The spectrum of any Pixel can be obtained through a cut over the lambda stack by plot- ting the intensity vs. wavelength. The wavelength selec- tion can be realized either by a rotating wheel with fixed band pass filters, acousto-optical-tunable filter (AOTF), - high lateral resolution liquid crystal tunable filters (LCTF) or by monochromatic - easy to implement illumination. This mode leads to a band-sequential im- aging (BSQ) data format. - high information density - stop motion method Pushbroom Imaging Pushbroom This system uses a matrix detector together with a spectrograph and a lens system. Images contain the full spectral information along one line across the sample.