Visualizing the Early Infection of Agathis Australis by Phytophthora Agathidicida, Using Microscopy and fluorescent in Situ Hybridization
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
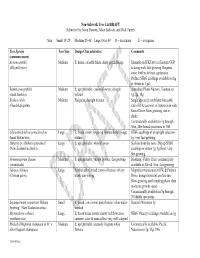
Recommended Non-Sidewalk Tree List DRAFT
Non-Sidewalk Tree List DRAFT Submitted by Jason Dewees, Mike Sullivan, and Dick Turner Size: Small 15-25’ Medium 25-40’ Large Over 40’ D = deciduous E = evergreen Tree Species Tree Size Design Characteristics Comments (common name) Acmena smithii Medium E; dense, colorful fruits; shiny green foliage Example on JFK Drive in Eastern GGP (lilly-pilly tree) is doing well; fast-growing. Requires some fertility for best appearance. Profuse SFBG seedlings available to dig or obtain in 1 gal. Banksia integrifolia Medium E; upright habit; creamy flowers; drought Australian Plants Nursery, Ventura, in (coast banksia) tolerant 1g, 5g, 15g Brahea edulis Medium Fan palm; drought tolerant Single specimen established on north (Guadalupe palm) side of JFK just west of intersection with Kezar Drive. Slow growing, sun or shade. Commercially available in 1g through 36in, 48in boxed specimens to 15ft Chiranthodendron pentadactylon Large E; broad crown; large red flowers, bold foliage SFBG seedlings of an upright selection: (hand flower tree) texture 1g; very fast-growing Hoheria sp. (Hoheria populnea? Large E; upright habit; white flowers Suckers from the roots. Dig up SFBG (New Zealand lacebark)) seedlings or obtain 1g, 5g there; very fast-growing Hymenosporum flavum Medium E; upright habit; yellow flowers; fast growing Boething, Valley Crest; commercially (sweetshade) available in 24in & 36in; fast-growing. Jubaea chilensis Large Feather palm; broad crown of leaves; silvery Magnificent specimen at JFK & Fuchsia (Chilean palm) trunk; sun-loving Drive; drought-tolerant, prefers sun. Slow-growing until trunking phase, then moderate growth speed. Commercially available in 5g through 20ft B&B specimens Leptospermum scoparium ‘Helene Small E; broad, low crown; pink flowers; often multi- Suncrest Nurseries 5g Strybing’ (New Zealand tea tree) trunked Metrosideros robusta Large E; broad dense crown; showy red flowers in SFBG Nursery seedlings available in 1g (northern rata) summer; slow & unusual but very well-adapted Michelia/Magnolia champaca or M. -

Agathis Robusta and Agathis Australis Friends Friends
Plants in Focus, December 2016 Agathis robusta and Agathis australis Friends of GeelongBotanic Left: The Qld Kauri Agathis robusta, planted in the Albury BG in 1910, is the largest recorded in the Big Tree Register. Note gardener. [1] Right: The NZ Kauri Agathis australis, named Tane Mahuta (Lord of the Forest), in the Waipoua Forest is the largest known in NZ. Photo: Prof. Chen Hualin, CC BY-SA 4.0, zh.wikipedia.org Kauris (Agathis sp.) are conifers Conifers, along with the other Gymnosperms (Cycads and Ginkgoes), first appeared about 300 Ma (Million years ago) at the end of the Carboniferous when the world’s coal deposits were being laid down with the remains of the spore-producing trees of that period. The early conifers looked like modern Araucaria. These trees spread throughout the world and displaced their predecessors. The age of the seed plants had arrived. The conifers are a hardy lot. They survived the largest mass extinction the earth has known, 252 Ma, at the end of the Permian Period. But more challenges lay ahead. Sometime in the next 50 Myr (Million years) one of Gymnosperms gave rise to the flowering plants, the Angiosperms. By 100 Ma, in the Cretaceous period, Angiosperms were widespread. And so the battle began - and still continues to this day. The flowering plants have many features that make them more successful in many environments, so their take-over of many habitats was complete by about 65 Ma at the end of the age of the dinosaurs. But in the world’s harsh environments the conifers continue to not just survive, but flourish. -

26 Extreme Trees Pub 2020
Publication WSFNR-20-22C April 2020 Extreme Trees: Tallest, Biggest, Oldest Dr. Kim D. Coder, Professor of Tree Biology & Health Care / University Hill Fellow University of Georgia Warnell School of Forestry & Natural Resources Trees have a long relationship with people. They are both utility and amenity. Trees can evoke awe, mysticism, and reverence. Trees represent great public and private values. Trees most noticed and celebrated by people and communities are the one-tenth of one-percent of trees which approach the limits of their maximum size, reach, extent, and age. These singular, historic, culturally significant, and massive extreme trees become symbols and icons of life on Earth, and our role model in environmental stewardship and sustainability. What Is A Tree? Figure 1 is a conglomeration of definitions and concepts about trees from legal and word definitions in North America. For example, 20 percent of all definitions specifically state a tree is a plant. Concentrated in 63% of all descriptors for trees are four terms: plant, woody, single stem, and tall. If broad stem diameter, branching, and perennial growth habit concepts are added, 87% of all the descriptors are represented. At its most basic level, defining a tree is not species based, but is a structural definition. A tree is represented by a type of plant architecture recognizable by non-technical people. The most basic concepts for defining a tree are — a large, tall, woody, perennial plant with a single, unbranched, erect, self-supporting stem holding an elevated and distinct crown of branches, and which is greater than 10 feet in height and greater than 3 inches in diameter. -

NOTES on AGATHIS AUSTRALIS. by C
NOTES ON AGATHIS AUSTRALIS. By C. T. SANDO. Introduction.—Much has been published about our kauri (Agathis australis) and it is now generally recognised as one of the finest coniferous trees of the world. The New Zealand species is one of a genus of ten—the most important being— A. Palmerstoni ... (Queensland) A. microstachya ... (Queensland) A. robusta ... (Queensland and Philippine Islands) A. vitiensis * ... (Fiji) A. alba ... (Malaya, Sumatra, Java, Celebes, Borneo, and Philippine Islands) A. lanceolata ... (New Caledonia) A. macrophylla ... (Solomon Islands) The value of Agathis australis was first recognised before 1800 by which time a considerable trade in kauri spars had sprung up between Australia and New Zealand. Later, naval-store ships were sent out from England for cargoes and from that time on kauri has increased in value. Up till a short time ago it was used for building and interior furnishing, but now the limited supplies demand a price far too high for these purposes so that its present uses have been confined to carriage building, cabinet-making, and general joinery, vats, tubs, and other special purposes. Wanton waste has so depleted out kauri areas that now, more than ever it is forced upon us, the necessity of making very serious attempts at conservation of the remaining supplies and propagation of this tree for scenic purposes, and, if economic conditions warrant it, for timber production. Geographical, Climatic, and Edaphic Range.—Kauri occurs in the northern parts of the North Island except in the extreme northern peninsula. It is found from Ahipara and Mangonui in the north as far south as the Bay of Plenty on the East Coast and Kawhia Harbour on the west, i.e. -

The Scientific Reserves of Auckland University. Ii. Quantitative Vegetation Studies
TANE 29, 1983 THE SCIENTIFIC RESERVES OF AUCKLAND UNIVERSITY. II. QUANTITATIVE VEGETATION STUDIES by John Ogden Department of Botany, University of Auckland, Private Bag, Auckland SUMMARY Forest vegetation surveys, using both plots and point-centred-quarter (plotless) methods, were undertaken by students in the University reserves at Swanson, Huapai, Oratia and Leigh over the period 1979 to 1982. The two methods gave similar estimates of species composition, total basal area and total density, but both gave wide confidence intervals on the parameters being estimated. The "leading species", in terms of both density and basal area, are defined for each area. The differences between the rankings for these different vegetation parameters are commented upon A regression of age on height of kauri (Agathis australis) seedlings is presented. The relationship between age and trunk diameter is discussed. It is shown that in cores taken from kauri poles in ricker stands over the size range 10 to 40 cm DBH the relationship between age and diameter is very weak. When a wider size range is employed a highly statistically significant relationship is obtained. It is concluded that the highly skewed size class frequency distribution of kauri, characteristic of such stands, reflects a competitive hierarchy. The senile population structure of kanuka (Leptospermum ericoides) in ricker stands confirms their serai role in forest development. The basal area estimates are discussed in the context of other figures for New Zealand forests. It is concluded that although the values are high (average 70 m2 ha 1) they are nevertheless below those commonly found in mature kauri forest. -

Hidden Fungi: Combining Culture-Dependent and -Independent DNA Barcoding Reveals Inter-Plant Variation in Species Richness of Endophytic Root Fungi in Elymus Repens
Journal of Fungi Article Hidden Fungi: Combining Culture-Dependent and -Independent DNA Barcoding Reveals Inter-Plant Variation in Species Richness of Endophytic Root Fungi in Elymus repens Anna K. Høyer and Trevor R. Hodkinson * Botany, School of Natural Sciences, Trinity College Dublin, The University of Dublin, Dublin D2, Ireland; [email protected] * Correspondence: [email protected] Abstract: The root endophyte community of the grass species Elymus repens was investigated using both a culture-dependent approach and a direct amplicon sequencing method across five sites and from individual plants. There was much heterogeneity across the five sites and among individual plants. Focusing on one site, 349 OTUs were identified by direct amplicon sequencing but only 66 OTUs were cultured. The two approaches shared ten OTUs and the majority of cultured endo- phytes do not overlap with the amplicon dataset. Media influenced the cultured species richness and without the inclusion of 2% MEA and full-strength MEA, approximately half of the unique OTUs would not have been isolated using only PDA. Combining both culture-dependent and -independent methods for the most accurate determination of root fungal species richness is therefore recom- mended. High inter-plant variation in fungal species richness was demonstrated, which highlights the need to rethink the scale at which we describe endophyte communities. Citation: Høyer, A.K.; Hodkinson, T.R. Hidden Fungi: Combining Culture-Dependent and -Independent Keywords: DNA barcoding; Elymus repens; fungal root endophytes; high-throughput amplicon DNA Barcoding Reveals Inter-Plant sequencing; MEA; PDA Variation in Species Richness of Endophytic Root Fungi in Elymus repens. J. Fungi 2021, 7, 466. -

Screening a Natural Product-Inspired Library for Anti-Phytophthora Activities
molecules Article Screening a Natural Product-Inspired Library for Anti-Phytophthora Activities Scott A. Lawrence 1, Hannah F. Robinson 2 , Daniel P. Furkert 3, Margaret A. Brimble 3 and Monica L. Gerth 2,* 1 Department of Microbiology and Immunology, University of Otago, Dunedin 9054, New Zealand; [email protected] 2 School of Biological Sciences, Victoria University of Wellington, Wellington 6140, New Zealand; [email protected] 3 School of Chemical Sciences, University of Auckland, Auckland 1142, New Zealand; [email protected] (D.P.F.); [email protected] (M.A.B.) * Correspondence: [email protected] Abstract: Phytophthora is a genus of microorganisms that cause devastating dieback and root-rot diseases in thousands of plant hosts worldwide. The economic impact of Phytophthora diseases on crops and native ecosystems is estimated to be billions of dollars per annum. These invasive pathogens are extremely difficult to control using existing chemical means, and the effectiveness of the few treatments available is being jeopardized by increasing rates of resistance. There is an urgent need to identify new chemical treatments that are effective against Phytophthora diseases. Natural products have long been regarded as “Nature’s medicine chest”, providing invaluable leads for developing front-line drugs and agrochemical agents. Here, we have screened a natural product- inspired library of 328 chemicals against two key Phytophthora species: Phytophthora cinnamomi and Phytophthora agathidicida. The library was initially screened for inhibition of zoospore germination. Citation: Lawrence, S.A.; Robinson, From these screens, we identified twenty-one hits that inhibited germination of one or both species. -

Kauri Care Guide About This Booklet This Booklet Is for Landowners, Managers and Occupiers with Kauri on Their Land
kauri care guide About this booklet This booklet is for landowners, managers and occupiers with kauri on their land. It explains the ecology of kauri, common diseases and gives advice about care and propagation. This booklet has three main sections: 1. Kauri ecology We describe the kauri family, how kauri grow and reproduce, and diseases that affect these trees. 2. How to care for your kauri We give advice on how to reduce stress and care for your kauri. 3. How to grow kauri We describe how to gather seed, propagate and plant out kauri. A glossary of terms used and list of further reading is also included. Acknowledgements Compiled by Stacey Hill, Biosecurity, Auckland Council. Content provided by Auckland Council: Nick Waipara, Plant Pathologist, Biosecurity; Amanda Peart, Communications Advisor, Biosecurity; Brendan Mackie, Horticulturist/ Ranger; Damon Keen, Creative Direction Advisor; Auckland Council Natural Heritage Team; Auckland Council Biosecurity Team. Thanks also to: Ian Barton, Forestry and Environmental Consultant; Ian Horner, Plant Pathologist, Plant and Food Research and John Hogan, Three Streams Reserve. ISBN No: 978-1-98-857134-8 (print) ISBN No: 978-1-98-857133-1 (online) Disclaimer While every effort has been made to ensure the information in this publication is accurate, the Kauri Dieback Programme does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Any view or opinion expressed -

Re-Evaluation of Forest Biomass Carbon Stocks and Lessons from the World’S Most Carbon-Dense Forests
Re-evaluation of forest biomass carbon stocks and lessons from the world’s most carbon-dense forests Heather Keith1, Brendan G. Mackey, and David B. Lindenmayer The Fenner School of Environment and Society, Australian National University, Canberra, ACT 0200, Australia Communicated by Gene E. Likens, Cary Institute of Ecosystem Studies, Millbrook, NY, March 9, 2009 (received for review July 14, 2008) from functioning as carbon sinks (11–13). The long time it takes new (136 ؍ From analysis of published global site biomass data (n primary forests, we discovered (i) the world’s highest known total plantings to sequester and store the amount of carbon equivalent to biomass carbon density (living plus dead) of 1,867 tonnes carbon per that stored in mature forests counters the second argument (14). ha (average value from 13 sites) occurs in Australian temperate moist The third argument about the unimportance of old forest in Eucalyptus regnans forests, and (ii) average values of the global site addressing climate change relates, in part, to the diminishing extent of primary forest caused by land-use activities (15) and associated ؍ biomass data were higher for sampled temperate moist forests (n forests (n depletion of biomass carbon stocks (16). However, significant areas (52 ؍ and boreal (n (36 ؍ than for sampled tropical (n (44 is number of sites per forest biome). Spatially averaged Intergovern- of primary forest remain (17), and depleted carbon stocks in mental Panel on Climate Change biome default values are lower than modified forests can be restored. our average site values for temperate moist forests, because the It is useful to distinguish between the carbon carrying capacity of temperate biome contains a diversity of forest ecosystem types that a forest ecosystem and its current carbon stock. -

A History of Kauri
A history of kauri Gretel Boswijk School of Geography and Environmental Science, The University of Auckland, New Zealand Keywords: Agathis australis, dendrochronology, kauri, New Zealand, palynology, tree-ring analysis Introduction The New Zealand kauri (Agathis australis) is a species notable for the height, bulk and longevity of the trees, and the good quality of its timber. Kauri towers above the canopy of the surrounding lowland forest (Figure 1), often occurring in stands of trees which are usually of a similar age. There are records of kauri attaining heights over 50 metres, girths greater than ten metres, and of trees which are estimated to be well over 1000 years old. These attributes of kauri—size, age, as well as timber quantity and quality—combine to make it an iconic tree which holds a special place in New Zealand landscape and culture. This is despite (or rather enhanced by) the natural distribution of kauri being restricted to the warm temperate forests of the upper North Island (Figure 2). Ferdinand Hochstetter, an Austrian scientist who visited New Zealand in the 1850s wrote that ‘Three degrees of longitude [173°–176° long] and three degrees of latitude [34½° - 37½° lat]…encompasses the entire and the only range of this remarkable tree’ (Hochstetter 1867: 141), although in the modern day, planted specimens can be found throughout the country. Kauri features in the Maori creation myth. When Tane, god and father of the forests and its creatures, separated his parents Rangi nui (Sky Father) and Papa-tu-a-nuku (Earth mother) to bring light and beauty into a dark world, he grew like a kauri. -

Phytochemistry, Chemotaxonomy, and Biological Activities of the Araucariaceae Family—A Review
plants Review Phytochemistry, Chemotaxonomy, and Biological Activities of the Araucariaceae Family—A Review Claudio Frezza 1,* , Alessandro Venditti 2 , Daniela De Vita 1, Chiara Toniolo 1, Marco Franceschin 2, Antonio Ventrone 1, Lamberto Tomassini 1 , Sebastiano Foddai 1, Marcella Guiso 2, Marcello Nicoletti 1, Armandodoriano Bianco 2 and Mauro Serafini 1 1 Dipartimento di Biologia Ambientale, Università di Roma “La Sapienza”, Piazzale Aldo Moro 5, 00185 Rome, Italy; [email protected] (D.D.V.); [email protected] (C.T.); [email protected] (A.V.); [email protected] (L.T.); [email protected] (S.F.); [email protected] (M.N.); mauro.serafi[email protected] (M.S.) 2 Dipartimento di Chimica, Università di Roma “La Sapienza”, Piazzale Aldo Moro 5, 00185 Rome, Italy; [email protected] (A.V.); [email protected] (M.F.); [email protected] (M.G.); [email protected] (A.B.) * Correspondence: [email protected] Received: 22 June 2020; Accepted: 9 July 2020; Published: 14 July 2020 Abstract: In this review article, the phytochemistry of the species belonging to the Araucariaceae family is explored. Among these, in particular, it is given a wide overview on the phytochemical profile of Wollemia genus, for the first time. In addition to this, the ethnopharmacology and the general biological activities associated to the Araucariaceae species are singularly described. Lastly, the chemotaxonomy at the genus and family levels is described and detailed. Keywords: Araucariaceae; phytochemistry; ethnopharmacology; chemotaxonomy; biological activities 1. Introduction Araucariaceae Henkel and W. Hochstetter is a family of coniferous trees, classified under the order Pinales, the class Pinopsoda, the division Pinophyta, and the Clade Tracheophytes [1]. -

Nutrition and Assessment of Current Nursery Container Mixes
350 Combined Proceedings International Plant Propagators' Society, Volume 43,1993 A Review of Kauri (Agathis australis) Nutrition and Assessment of Current Nursery Container Mixes Michael B. Thomas and Mervyn I. Spurway Horticulture Department, Lincoln University, Canterbury INTRODUCTION Kauri forests once covered much of the early land mass of New Zealand. Now only a remnant of the great forests remain-a mere 5% of the pre-European kauri forest has been spared from the axe and burning (Ahmed and Ogden, 1987). Kauri is a large, long-lived tree which normally grows for more than 600 years with individuals exceeding 2 m d.b.h. (diameter at breast height) and often 1000 years ofage (Ahmed and Ogden, 1987). Sale (1978) in his book, which reviews the history ofthe kauri in New Zealand, states that forest authorities consider this to be the tree capable ofyielding the greatest volume ofmillable timber in the world. Sale goes on to explain the significance ofthis tree in the early maori history and European colonisation, includingits uses and the wastage thathas occurred. Fires were particularly common in logged kauri forests, many being deliberately lit by gum diggers (Ecroyd, 1982). Fortunately some great kauri giants have remained and are protected in reserves and parks. Many are seen as historic and notable trees and have been individually named (Sale, 1978; Burstall and Sale, 1984; Salmon, 1990). The kauri is therefore seen as having great historic and amenity value in New Zealand. Recently there has been a new interest in it as a forestry tree. Early papers have recorded how the kauri is readily produced from seed (Morrison, 1955; Dakin and McClure, 1975; Barton, 1978) and also by vegetative propagation (Dakin and Mearns, 1975).