Model-Driven Architecture for Cancer Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Eves2005-Dformstfp04
Chapter 1 Disjoint Forms in Graphical User Interfaces Sander Evers, Peter Achten, Rinus Plasmeijer1 Abstract: Forms are parts of a graphical user interface (GUI) that show a (struc- tured) value and allow the user to update it. Some forms express a choice between two or more (structured) values using radio buttons or check boxes. We show that explicitly modelling such a choice leads to a cleaner separation of logic and lay- out. This is done by extending the combinator library FunctionalForms with dis- joint form combinators. To implement these, we have generalized the technique of compositional functional references which underlies the library. 1.1 INTRODUCTION Forms are parts of a graphical user interface (GUI) that show a (structured) value and allow the user to update it. For example, the omnipresent dialogs labeled Options, Settings and Preferences are forms. An address book can also be con- sidered a form. In our previous work, we have developed the combinator library FunctionalForms[2] for building forms in a concise, compositional way. Many real-life forms allow a choice between two or more alternatives, some of which require extra information. For example, the form in Fig. 1.1 indicates whether the user wishes to receive a certain newsletter; if s/he does, the text entry field next to this option should contain his/her email adress. If s/he does not, this text field is irrelevant (some GUIs provide a visual clue for this: the control is dimmed). Usually, the information in such a form is processed as a product-like data structure containing the choice (e.g. -

Comparison of Common Xml-Based Web User Interface Languages
Journal of Web Engineering, Vol. 9, No. 2 (2010) 095–115 c Rinton Press COMPARISON OF COMMON XML-BASED WEB USER INTERFACE LANGUAGES MIKKO POHJA Department of Media Technology, Aalto University P.O. Box 15400, FI-00076 Aalto, Finland mikko.pohja@hut.fi Received August 1, 2009 Revised February 25, 2010 In addition to being a platform for information access, the World Wide Web is increas- ingly becoming an application platform. While web applications have several benefits compared to desktop applications, there are also some problems. With legacy HTML, for example, one cannot produce user interfaces such as those that users have become accustomed to with desktop applications. What worked for static documents is not suf- ficient for the complicated web applications of today. Several parties have addressed this problem by defining a specific UI description language. In addition, the renewal of HTML aims to enhance support for web applications. This study evaluated five XML- based UI description formats, including HTML 5, in order to determine which language is best suited for modern web application development. The study also assessed what kind of applications are suited to each format. The requirements for a Web UI descrip- tion language from the literature were revised and three use cases were defined, through which the languages are evaluated. The paper also presents the model differences of the languages. Keywords: Web User Interface Description Language, Web Application Communicated by: D. Lowe & O. Pastor 1 Introduction Commerce and communication tasks, such as the use of e-mail, are common today on the World Wide Web (WWW), as is a trend towards realizing higher interaction tasks, such as in- formation authoring. -

Microsoft, Adobe & W3C to Shake up Electronic Forms Market
Vol. 11, No, 8 October 2003 www.gilbane.com Published by: Bluebill Advisors, Inc. 763 Massachusetts Ave. Cambridge, MA 02139 USA (617) 497.9443 Fax (617) 497.5256 www.bluebilladvisors.com Editor: Frank Gilbane [email protected] (617) 497.9443 Content Technology Works! Editors Emeriti: Tim Bray [email protected] (604) 708.9592 MICROSOFT, ADOBE & XFORMS TO David Weinberger [email protected] (617) 738.8323 SHAKE UP ELECTRONIC FORMS MARKET Senior Editors: Sebastian Holst [email protected] Our title this month reads like a news headline on purpose. There are a number Bill Trippe [email protected] of new, and upcoming, developments in electronic forms (eForms) technology (617) 497.9443 that should be grabbing your attention. Some of these are of major importance Recent Contributors: on their own, but taken together, they signal the start of a major improvement Kathleen Reidy in businesses’ ability to easily collect, integrate, and process information. [email protected] Bob Doyle [email protected] “Electronic forms” have been around for years, but the term refers to a wide variety of technologies – from scanned image applications to HTML forms – Production Assistant: Sarah G. Dionne that are not at all similar and far from equal in their ability to accelerate and [email protected] smooth business processes. What eForm technology has shared is: a level of (617) 497.9443 difficulty that kept it out of the reach of office professionals who were com- Subscriptions: fortable enough with documents and spreadsheets, but scared-off by forms, [email protected] (617) 497.9443 and proprietary data formats that made information integration costly and complex. -

A Declarative Approach Based on Xforms
Helsinki University of Technology Publications in Telecommunications Software and Multimedia Teknillisen korkeakoulun tietoliikenneohjelmistojen ja multimedian julkaisuja Espoo 2006 TML-A16 WEB USER INTERACTION - A DECLARATIVE APPROACH BASED ON XFORMS Mikko Honkala Dissertation for the degree of Doctor of Science in Technology to be presented with due permission of the Department of Computer Science and Engineering, for pub- lic examination and debate in Auditorium T2 at Helsinki University of Technology (Espoo, Finland) on the 12th of January, 2007, at 12 noon. Helsinki University of Technology Department of Computer Science and Engineering Telecommunications Software and Multimedia Laboratory Teknillinen korkeakoulu Tietotekniikan osasto Tietoliikenneohjelmistojen ja multimedian laboratorio Distribution: Helsinki University of Technology Telecommunications Software and Multimedia Laboratory P.O.Box 5400 FIN-02015 HUT Tel. +358-9-451 2870 Fax. +358-9-451 5014 c Mikko Honkala ISBN-13 978-951-22-8565-5 ISBN-10 951-22-8565-7 ISSN 1456-7911 ISBN-13 978-951-22-8566-2 (PDF) ISBN-10 951-22-8566-5 (PDF) ISSN 1455 9722 (PDF) URL: http://lib.tkk.fi/Diss/ Otamedia Oy Espoo 2006 ABSTRACT Author Mikko Honkala Title Web User Interaction - a Declarative Approach Based on XForms Published Doctoral thesis, Helsinki University of Technology, 2006 Keywords XML, User Interfaces, User Interaction, XForms, UIDL, XHTML This thesis studies next-generation web user interaction definition languages, as well as browser software architectures. The motivation comes from new end-user requirements for web applications: demand for higher interaction, adaptation for mobile and multimodal usage, and rich multimedia content. At the same time, there is a requirement for non- programmers to be able to author, customize, and maintain web user interfaces. -

Multiplatformní GUI Toolkity GTK+ a Qt
Multiplatformní GUI toolkity GTK+ a Qt Jan Outrata KATEDRA INFORMATIKY UNIVERZITA PALACKÉHO V OLOMOUCI GUI toolkit (widget toolkit) (1) = programová knihovna (nebo kolekce knihoven) implementující prvky GUI = widgety (tlačítka, seznamy, menu, posuvník, bary, dialog, okno atd.) a umožňující tvorbu GUI (grafického uživatelského rozhraní) aplikace vlastní jednotný nebo nativní (pro platformu/systém) vzhled widgetů, možnost stylování nízkoúrovňové (Xt a Xlib v X Windows System a libwayland ve Waylandu na unixových systémech, GDI Windows API, Quartz a Carbon v Apple Mac OS) a vysokoúrovňové (MFC, WTL, WPF a Windows Forms v MS Windows, Cocoa v Apple Mac OS X, Motif/Lesstif, Xaw a XForms na unixových systémech) multiplatformní = pro více platforem (MS Windows, GNU/Linux, Apple Mac OS X, mobilní) nebo platformově nezávislé (Java) – aplikace může být také (většinou) událostmi řízené programování (event-driven programming) – toolkit v hlavní smyčce zachytává události (uživatelské od myši nebo klávesnice, od časovače, systému, aplikace samotné atd.) a umožňuje implementaci vlastních obsluh (even handler, callback function), objektově orientované programování (objekty = widgety aj.) – nevyžaduje OO programovací jazyk! Jan Outrata (Univerzita Palackého v Olomouci) Multiplatformní GUI toolkity duben 2015 1 / 10 GUI toolkit (widget toolkit) (2) language binding = API (aplikační programové rozhraní) toolkitu v jiném prog. jazyce než původní API a toolkit samotný GUI designer/builder = WYSIWYG nástroj pro tvorbu GUI s využitím toolkitu, hierarchicky skládáním prvků, z uloženého XML pak generuje kód nebo GUI vytvoří za běhu aplikace nekomerční (GNU (L)GPL, MIT, open source) i komerční licence např. GTK+ (C), Qt (C++), wxWidgets (C++), FLTK (C++), CEGUI (C++), Swing/JFC (Java), SWT (Java), JavaFX (Java), Tcl/Tk (Tcl), XUL (XML) aj. -
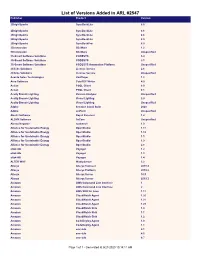
List of Versions Added in ARL #2547 Publisher Product Version
List of Versions Added in ARL #2547 Publisher Product Version 2BrightSparks SyncBackLite 8.5 2BrightSparks SyncBackLite 8.6 2BrightSparks SyncBackLite 8.8 2BrightSparks SyncBackLite 8.9 2BrightSparks SyncBackPro 5.9 3Dconnexion 3DxWare 1.2 3Dconnexion 3DxWare Unspecified 3S-Smart Software Solutions CODESYS 3.4 3S-Smart Software Solutions CODESYS 3.5 3S-Smart Software Solutions CODESYS Automation Platform Unspecified 4Clicks Solutions License Service 2.6 4Clicks Solutions License Service Unspecified Acarda Sales Technologies VoxPlayer 1.2 Acro Software CutePDF Writer 4.0 Actian PSQL Client 8.0 Actian PSQL Client 8.1 Acuity Brands Lighting Version Analyzer Unspecified Acuity Brands Lighting Visual Lighting 2.0 Acuity Brands Lighting Visual Lighting Unspecified Adobe Creative Cloud Suite 2020 Adobe JetForm Unspecified Alastri Software Rapid Reserver 1.4 ALDYN Software SvCom Unspecified Alexey Kopytov sysbench 1.0 Alliance for Sustainable Energy OpenStudio 1.11 Alliance for Sustainable Energy OpenStudio 1.12 Alliance for Sustainable Energy OpenStudio 1.5 Alliance for Sustainable Energy OpenStudio 1.9 Alliance for Sustainable Energy OpenStudio 2.8 alta4 AG Voyager 1.2 alta4 AG Voyager 1.3 alta4 AG Voyager 1.4 ALTER WAY WampServer 3.2 Alteryx Alteryx Connect 2019.4 Alteryx Alteryx Platform 2019.2 Alteryx Alteryx Server 10.5 Alteryx Alteryx Server 2019.3 Amazon AWS Command Line Interface 1 Amazon AWS Command Line Interface 2 Amazon AWS SDK for Java 1.11 Amazon CloudWatch Agent 1.20 Amazon CloudWatch Agent 1.21 Amazon CloudWatch Agent 1.23 Amazon -

An MVC-Based Intelligent Document Model Using UIML
Information 2015, 6, 122-133; doi:10.3390/info6020122 OPEN ACCESS information ISSN 2078-2489 www.mdpi.com/journal/information Article An MVC-based Intelligent Document Model Using UIML Yunmei Shi *, Xuhong Liu †, Ning Li † and Xia Hou Computer School, Beijing Information Science & Technology University, No 35 Beisihuan Zhonglu, Chaoyang District, Beijing 100101, China; E-Mails: [email protected] (X.L.); [email protected] (N.L.); [email protected] (X.H.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel./Fax: +86-10-6488-4434. Academic Editor: Willy Susilo Received: 29 December 2014 / Accepted: 11 March 2015 /Published: 27 March 2015 Abstract: Aiming at the common problems of intelligent document platform-dependency, this paper proposes an MVC-based (Model View Controller-based) intelligent document model using UIML (User Interface Markup Language). The model is made on the basis of the previous work of our team, and the difference is that the new model separates user interface and interaction descriptions from the view component to make the intelligent document model much more independent of platform and programming language. To verify the intelligent document model, we implemented a prototype, which can support intelligent operations. The test result shows that our approach is correct. The model not only follows MVC framework, but also provides good flexibility and independence. Keywords: intelligent document; user interface; UIML; MVC 1. Introduction Traditionally, a document can only display content by office software according to the static description information of the document. These kinds of documents are usually called “static documents”, whose characteristic is lacking of interactivity and operability. -

Pipenightdreams Osgcal-Doc Mumudvb Mpg123-Alsa Tbb
pipenightdreams osgcal-doc mumudvb mpg123-alsa tbb-examples libgammu4-dbg gcc-4.1-doc snort-rules-default davical cutmp3 libevolution5.0-cil aspell-am python-gobject-doc openoffice.org-l10n-mn libc6-xen xserver-xorg trophy-data t38modem pioneers-console libnb-platform10-java libgtkglext1-ruby libboost-wave1.39-dev drgenius bfbtester libchromexvmcpro1 isdnutils-xtools ubuntuone-client openoffice.org2-math openoffice.org-l10n-lt lsb-cxx-ia32 kdeartwork-emoticons-kde4 wmpuzzle trafshow python-plplot lx-gdb link-monitor-applet libscm-dev liblog-agent-logger-perl libccrtp-doc libclass-throwable-perl kde-i18n-csb jack-jconv hamradio-menus coinor-libvol-doc msx-emulator bitbake nabi language-pack-gnome-zh libpaperg popularity-contest xracer-tools xfont-nexus opendrim-lmp-baseserver libvorbisfile-ruby liblinebreak-doc libgfcui-2.0-0c2a-dbg libblacs-mpi-dev dict-freedict-spa-eng blender-ogrexml aspell-da x11-apps openoffice.org-l10n-lv openoffice.org-l10n-nl pnmtopng libodbcinstq1 libhsqldb-java-doc libmono-addins-gui0.2-cil sg3-utils linux-backports-modules-alsa-2.6.31-19-generic yorick-yeti-gsl python-pymssql plasma-widget-cpuload mcpp gpsim-lcd cl-csv libhtml-clean-perl asterisk-dbg apt-dater-dbg libgnome-mag1-dev language-pack-gnome-yo python-crypto svn-autoreleasedeb sugar-terminal-activity mii-diag maria-doc libplexus-component-api-java-doc libhugs-hgl-bundled libchipcard-libgwenhywfar47-plugins libghc6-random-dev freefem3d ezmlm cakephp-scripts aspell-ar ara-byte not+sparc openoffice.org-l10n-nn linux-backports-modules-karmic-generic-pae -

X Window System Architecture Overview HOWTO
X Window System Architecture Overview HOWTO Daniel Manrique [email protected] Revision History Revision 1.0.1 2001−05−22 Revised by: dm Some grammatical corrections, pointed out by Bill Staehle Revision 1.0 2001−05−20 Revised by: dm Initial LDP release. This document provides an overview of the X Window System's architecture, give a better understanding of its design, which components integrate with X and fit together to provide a working graphical environment and what choices are there regarding such components as window managers, toolkits and widget libraries, and desktop environments. X Window System Architecture Overview HOWTO Table of Contents 1. Preface..............................................................................................................................................................1 2. Introduction.....................................................................................................................................................2 3. The X Window System Architecture: overview...........................................................................................3 4. Window Managers..........................................................................................................................................4 5. Client Applications..........................................................................................................................................5 6. Widget Libraries or toolkits...........................................................................................................................6 -

Debian Build Dependecies
Debian build dependecies Valtteri Rahkonen valtteri.rahkonen@movial.fi Debian build dependecies by Valtteri Rahkonen Revision history Version: Author: Description: 2004-04-29 Rahkonen Changed title 2004-04-20 Rahkonen Added introduction, reorganized document 2004-04-19 Rahkonen Initial version Table of Contents 1. Introduction............................................................................................................................................1 2. Required source packages.....................................................................................................................2 2.1. Build dependencies .....................................................................................................................2 2.2. Misc packages .............................................................................................................................4 3. Optional source packages......................................................................................................................5 3.1. Build dependencies .....................................................................................................................5 3.2. Misc packages ...........................................................................................................................19 A. Required sources library dependencies ............................................................................................21 B. Optional sources library dependencies .............................................................................................22 -

Slobodan Softver U Obrazovanju
I SVEUČILIŠTE U ZAGREBU UČITELJSKI FAKULTET Slobodan softver u obrazovanju Predrag Oreški Vladimir Šimović Zagreb, 2013. II Izdavač Učiteljski fakultet Sveučilišta u Zagrebu Savska cesta 77, Zagreb Za izdavača dr. sc. Ivan Prskalo, redoviti profesor Urednik dr. sc. Vladimir Šimović, redoviti profesor Recenzenti dr. sc. Nenad Prelog, redoviti profesor dr. sc. Ljubica Bakić Tomić, izvanredni profesor dr. sc. Dragutin Kermek, izvanredni profesor Lektorica Zdenka Župan Milković, prof. Ilustracija na naslovnici: Vector Open Stock Team, http://www.vectoropenstock.com/963-Vector-abstract-tree-vector Attribution No Derivatives 3.0 Unported License Prijelom: Nataša Rogulja Naklada 200 tiskanih primjeraka + online izdanje UDK 004.4.057.8:37 373.3:004 ISBN 978-953-7210-63-2 Tisak dovršen u 2013. Ovo djelo je dano na korištenje pod licencijom Creative Commons Imenovanje-Nekomercijalno-Bez prerada 3.0 nelokalizirana licencija. http://creativecommons.org/licenses/by-nc-nd/3.0/deed.hr III IV Sadržaj Predgovor ....................................................................................................................... VII Sažetak ......................................................................................................................... VIII 1. Uvod .............................................................................................................................. 1 2. Intelektualno vlasništvo ................................................................................................ 8 2.1. Intelektualno vlasništvo -

Improving General Practice Based Epidemiologic Surveillance Using Desktop Clients: the French Sentinel Network Experience
442 MEDINFO 2010 C. Safran et al. (Eds.) IOS Press, 2010 © 2010 IMIA and SAHIA. All rights reserved. doi:10.3233/978-1-60750-588-4-442 Improving general practice based epidemiologic surveillance using desktop clients: the French Sentinel Network experience a,b b,c Clément Turbelin and Pierre-Yves Boëlle a UPMC Univ Paris 06, UMR S 707, F-75012 Paris, France b INSERM, U707, F-75012 Paris, France c AP-HP, Hôpital Saint-Antoine, Paris, F-75012, France Abstract Estimates of disease incidence are computed from GPs reports and used for public health decision making. Introduction: Web-based applications are a choice tool for general practice based epidemiological surveillance; however The satisfaction surveys of GPs participating in the their use may disrupt the general practitioners (GPs) work surveillance alerted us to the fact that interaction with the process. In this article, we propose an alternative approach website could be a limitation to the doctors’ persistence in based on a desktop client application. This was developed for surveillance. Indeed, with this system, GPs must periodically use in the French General Practitioners Sentinel Network. log in to a secure web site and report cases using a dedicated Methods: We developed a java application running as a client interface (see Figure 1). This process is difficult to perform on the local GP computer. It allows reporting cases to a during consultation, and leads to delays in reporting. As public central server and provides feedback to the participating GPs. health decision makers are increasingly expecting near real XML was used to describe surveillance protocols and time reporting and analysis of data [3-4], it is relevant to try questionnaires as well as instances of case descriptions.