Are We There Yet? Tracking the Development of New Model Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lineage-Specific Mapping of Quantitative Trait Loci
Heredity (2013) 111, 106–113 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Lineage-specific mapping of quantitative trait loci C Chen1 and K Ritland We present an approach for quantitative trait locus (QTL) mapping, termed as ‘lineage-specific QTL mapping’, for inferring allelic changes of QTL evolution along with branches in a phylogeny. We describe and analyze the simplest case: by adding a third taxon into the normal procedure of QTL mapping between pairs of taxa, such inferences can be made along lineages to a presumed common ancestor. Although comparisons of QTL maps among species can identify homology of QTLs by apparent co- location, lineage-specific mapping of QTL can classify homology into (1) orthology (shared origin of QTL) versus (2) paralogy (independent origin of QTL within resolution of map distance). In this light, we present a graphical method that identifies six modes of QTL evolution in a three taxon comparison. We then apply our model to map lineage-specific QTLs for inbreeding among three taxa of yellow monkey-flower: Mimulus guttatus and two inbreeders M. platycalyx and M. micranthus, but critically assuming outcrossing was the ancestral state. The two most common modes of homology across traits were orthologous (shared ancestry of mutation for QTL alleles). The outbreeder M. guttatus had the fewest lineage-specific QTL, in accordance with the presumed ancestry of outbreeding. Extensions of lineage-specific QTL mapping to other types of data and crosses, and to inference of ancestral QTL state, are discussed. Heredity (2013) 111, 106–113; doi:10.1038/hdy.2013.24; published online 24 April 2013 Keywords: Quantitative trait locus mapping; evolution of mating systems; Mimulus INTRODUCTION common ancestor of these two taxa, back to the common ancestor of Knowledge of the genetic architecture of complex traits can offer all three taxa and forwards again to the third taxon. -
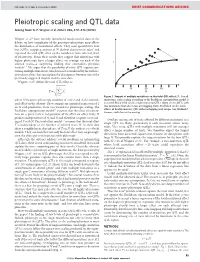
Pleiotropic Scaling and QTL Data Arising From: G
NATURE | Vol 456 | 4 December 2008 BRIEF COMMUNICATIONS ARISING Pleiotropic scaling and QTL data Arising from: G. P. Wagner et al. Nature 452, 470–473 (2008) Wagner et al.1 have recently introduced much-needed data to the debate on how complexity of the genotype–phenotype map affects the distribution of mutational effects. They used quantitative trait loci (QTLs) mapping analysis of 70 skeletal characters in mice2 and regressed the total QTL effect on the number of traits affected (level of pleiotropy). From their results they suggest that mutations with higher pleiotropy have a larger effect, on average, on each of the T affected traits—a surprising finding that contradicts previous models3–7. We argue that the possibility of some QTL regions con- taining multiple mutations, which was not considered by the authors, introduces a bias that can explain the discrepancy between one of the previously suggested models and the new data. Wagner et al.1 define the total QTL effect as vffiffiffiffiffiffiffiffiffiffiffiffiffi u uXN 10 20 30 ~t 2 T Ai N i~1 Figure 1 | Impact of multiple mutations on the total QTL effect, T. For all where N measures pleiotropy (number of traits) and Ai the normal- mutations, strict scaling according to the Euclidean superposition model is ized effect on the ith trait. They compare an empirical regression of T assumed. Black filled circles, single-mutation QTLs. Open circles, QTLs with on N with predictions from two models for pleiotropic scaling. The two mutations that affect non-overlapping traits. Red filled circles, total Euclidean superposition model3,4 assumes that the effect of a muta- effects of double-mutant QTL with overlapping trait ranges (see Methods). -

How Morphological Development Can Guide Evolution Sam Kriegman1,*, Nick Cheney1, and Josh Bongard1
How morphological development can guide evolution Sam Kriegman1,*, Nick Cheney1, and Josh Bongard1 1University of Vermont, Department of Computer Science, Burlington, VT, USA *[email protected] ABSTRACT Organisms result from adaptive processes interacting across different time scales. One such interaction is that between development and evolution. Models have shown that development sweeps over several traits in a single agent, sometimes exposing promising static traits. Subsequent evolution can then canalize these rare traits. Thus, development can, under the right conditions, increase evolvability. Here, we report on a previously unknown phenomenon when embodied agents are allowed to develop and evolve: Evolution discovers body plans robust to control changes, these body plans become genetically assimilated, yet controllers for these agents are not assimilated. This allows evolution to continue climbing fitness gradients by tinkering with the developmental programs for controllers within these permissive body plans. This exposes a previously unknown detail about the Baldwin effect: instead of all useful traits becoming genetically assimilated, only traits that render the agent robust to changes in other traits become assimilated. We refer to this as differential canalization. This finding also has implications for the evolutionary design of artificial and embodied agents such as robots: robots robust to internal changes in their controllers may also be robust to external changes in their environment, such as transferal from simulation to reality or deployment in novel environments. Introduction The shape of life changes on many different time scales. From generation to generation, populations gradually increase in complexity and relative competency. At the individual level, organisms grow from a single-celled egg and exhibit extreme postnatal change as they interact with the outside world during their lifetimes. -

The Polyp and the Medusa Life on the Move
The Polyp and the Medusa Life on the Move Millions of years ago, unlikely pioneers sparked a revolution. Cnidarians set animal life in motion. So much of what we take for granted today began with Cnidarians. FROM SHAPE OF LIFE The Polyp and the Medusa Life on the Move Take a moment to follow these instructions: Raise your right hand in front of your eyes. Make a fist. Make the peace sign with your first and second fingers. Make a fist again. Open your hand. Read the next paragraph. What you just did was exhibit a trait we associate with all animals, a trait called, quite simply, movement. And not only did you just move your hand, but you moved it after passing the idea of movement through your brain and nerve cells to command the muscles in your hand to obey. To do this, your body needs muscles to move and nerves to transmit and coordinate movement, whether voluntary or involuntary. The bit of business involved in making fists and peace signs is pretty complex behavior, but it pales by comparison with the suites of thought and movement associated with throwing a curve ball, walking, swimming, dancing, breathing, landing an airplane, running down prey, or fleeing a predator. But whether by thought or instinct, you and all animals except sponges have the ability to move and to carry out complex sequences of movement called behavior. In fact, movement is such a basic part of being an animal that we tend to define animalness as having the ability to move and behave. -

1 ROBERT PLOMIN on Behavioral Genetics Genetic Links Common
Genetic links • Background – Why has educational psychology ben so slow ROBERT PLOMIN to accept genetics? on Behavioral Genetics • Beyond nature vs. nurture – 3 findings from genetic research with far- reaching implications for education: October, 2004 The nature of nurture The abnormal is normal Generalist genes • Molecular Genetics Quantitative genetics: twin and adoption studies Common medical disorders Identical twins Fraternal twins (monozygotic, MZ) (dizygotic, DZ) 1 Twin concordances in TEDS at 7 years Twins Early Development Study (TEDS) for low reading and maths Perceptions of nature/nurture: % parents (N=1340) and teachers Beyond nature vs. nurture: (N=667) indicating that genes are at least as important as environment (Walker & Plomin, Ed. Psych., in press) The nature of nurture • Genetic influence on experience • Genotype-environment correlation – Children’s genetic propensities are correlated with their experiences passive evocative active 2 Beyond nature vs. nurture: Genotype-environment correlation The nature of nurture • Passive • Genotype-environment correlation – e.g., family background (SES) and school – Children’s genetic propensities are correlated with achievement their experiences passive • Evocative evocative – Children evoke experiences for genetic reasons Active • Active • Molecular genetics – Children actively create environments that foster – Find genes associated with learning experiences their genetic propensities Beyond nature vs. nurture: The one-gene one-disability (OGOD) model The abnormal is normal • Common learning disabilities are the quantitative extreme of the same genetic and environmental factors responsible for normal variation in learning abilities. • There are no common learning disabilities, just the extremes of normal variation. 3 OGOD (rare) and QTL (common) disorders The quantitative trait locus (QTL) model Beyond nature vs. -

Animal Origins and the Evolution of Body Plans 621
Animal Origins and the Evolution 32 of Body Plans In 1822, nearly forty years before Darwin wrote The Origin of Species, a French naturalist, Étienne Geoffroy Saint-Hilaire, was examining a lob- ster. He noticed that when he turned the lobster upside down and viewed it with its ventral surface up, its central nervous system was located above its digestive tract, which in turn was located above its heart—the same relative positions these systems have in mammals when viewed dorsally. His observations led Geoffroy to conclude that the differences between arthropods (such as lobsters) and vertebrates (such as mammals) could be explained if the embryos of one of those groups were inverted during development. Geoffroy’s suggestion was regarded as preposterous at the time and was largely dismissed until recently. However, the discovery of two genes that influence a sys- tem of extracellular signals involved in development has lent new support to Geof- froy’s seemingly outrageous hypothesis. Genes that Control Development A A vertebrate gene called chordin helps to establish cells on one side of the embryo human and a lobster carry similar genes that control the development of the body as dorsal and on the other as ventral. A probably homologous gene in fruit flies, called axis, but these genes position their body sog, acts in a similar manner, but has the opposite effect. Fly cells where sog is active systems inversely. A lobster’s nervous sys- become ventral, whereas vertebrate cells where chordin is active become dorsal. How- tem runs up its ventral (belly) surface, whereas a vertebrate’s runs down its dorsal ever, when sog mRNA is injected into an embryo (back) surface. -

Giraffe Stature and Neck Elongation: Vigilance As an Evolutionary Mechanism
biology Review Giraffe Stature and Neck Elongation: Vigilance as an Evolutionary Mechanism Edgar M. Williams Faculty of Life Sciences and Education, University of South Wales, Wales CF37 1DL, UK; [email protected]; Tel.: +44-1443-483-893 Academic Editor: Chris O’Callaghan Received: 1 August 2016; Accepted: 7 September 2016; Published: 12 September 2016 Abstract: Giraffe (Giraffa camelopardalis), with their long neck and legs, are unique amongst mammals. How these features evolved is a matter of conjecture. The two leading ideas are the high browse and the sexual-selection hypotheses. While both explain many of the characteristics and the behaviour of giraffe, neither is fully supported by the available evidence. The extended viewing horizon afforded by increased height and a need to maintain horizon vigilance, as a mechanism favouring the evolution of increased height is reviewed. In giraffe, vigilance of predators whilst feeding and drinking are important survival factors, as is the ability to interact with immediate herd members, young and male suitors. The evidence regarding giraffe vigilance behaviour is sparse and suggests that over-vigilance has a negative cost, serving as a distraction to feeding. In woodland savannah, increased height allows giraffe to see further, allowing each giraffe to increase the distance between its neighbours while browsing. Increased height allows the giraffe to see the early approach of predators, as well as bull males. It is postulated that the wider panorama afforded by an increase in height and longer neck has improved survival via allowing giraffe to browse safely over wider areas, decreasing competition within groups and with other herbivores. -

Hispid Pocket Mouse Chaetodipus Hispidus
Wyoming Species Account Hispid Pocket Mouse Chaetodipus hispidus REGULATORY STATUS USFWS: No special status USFS R2: No special status USFS R4: No special status Wyoming BLM: No special status State of Wyoming: Nongame Wildlife CONSERVATION RANKS USFWS: No special status WGFD: NSSU (U), Tier III WYNDD: G5, S1S3 Wyoming Contribution: LOW IUCN: Least Concern STATUS AND RANK COMMENTS The Wyoming Natural Diversity Database has assigned Hispid Pocket Mouse (Chaetodipus hispidus) a state conservation rank ranging from S1 (Critically Imperiled) to S3 (Vulnerable) because of uncertainty about the proportion of range occupied and population trends for this species in Wyoming. NATURAL HISTORY Taxonomy: Historically, there were four recognized subspecies of Hispid Pocket Mouse, and only C. h. paradoxus was found in Wyoming 1-5. A recent DNA-based study determined that the previously accepted subspecies are neither morphologically nor genetically distinct and instead proposed new subspecies boundaries delineated by four geographically and ecologically disjunct mitochondrial clades 6. Following this taxonomic revision, Wyoming remains within the distribution of the newly defined subspecies C. h. paradoxus 6. Description: It is possible to identify Hispid Pocket Mouse in the field. It is the largest Wyoming pocket mouse species; adults weigh between 40–60 g and can reach total lengths of 200–223 mm 2. Tail, hind foot, and ear length ranges from 90–113 mm, 25–28 mm, and 12–13 mm, respectively 2. Hispid Pocket Mouse is named for its distinctly coarse dorsal pelage, which is buff to yellowish orange mixed with black hairs, thus leading to an overall brownish or even olive appearance 1, 2, 4, 5, 7. -

The Cambrian Explosion: a Big Bang in the Evolution of Animals
The Cambrian Explosion A Big Bang in the Evolution of Animals Very suddenly, and at about the same horizon the world over, life showed up in the rocks with a bang. For most of Earth’s early history, there simply was no fossil record. Only recently have we come to discover otherwise: Life is virtually as old as the planet itself, and even the most ancient sedimentary rocks have yielded fossilized remains of primitive forms of life. NILES ELDREDGE, LIFE PULSE, EPISODES FROM THE STORY OF THE FOSSIL RECORD The Cambrian Explosion: A Big Bang in the Evolution of Animals Our home planet coalesced into a sphere about four-and-a-half-billion years ago, acquired water and carbon about four billion years ago, and less than a billion years later, according to microscopic fossils, organic cells began to show up in that inert matter. Single-celled life had begun. Single cells dominated life on the planet for billions of years before multicellular animals appeared. Fossils from 635,000 million years ago reveal fats that today are only produced by sponges. These biomarkers may be the earliest evidence of multi-cellular animals. Soon after we can see the shadowy impressions of more complex fans and jellies and things with no names that show that animal life was in an experimental phase (called the Ediacran period). Then suddenly, in the relatively short span of about twenty million years (given the usual pace of geologic time), life exploded in a radiation of abundance and diversity that contained the body plans of almost all the animals we know today. -

Epigenome-Wide Study Identified Methylation Sites Associated With
nutrients Article Epigenome-Wide Study Identified Methylation Sites Associated with the Risk of Obesity Majid Nikpay 1,*, Sepehr Ravati 2, Robert Dent 3 and Ruth McPherson 1,4,* 1 Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, 40 Ruskin St–H4208, Ottawa, ON K1Y 4W7, Canada 2 Plastenor Technologies Company, Montreal, QC H2P 2G4, Canada; [email protected] 3 Department of Medicine, Division of Endocrinology, University of Ottawa, the Ottawa Hospital, Ottawa, ON K1Y 4E9, Canada; [email protected] 4 Atherogenomics Laboratory, University of Ottawa Heart Institute, Ottawa, ON K1Y 4W7, Canada * Correspondence: [email protected] (M.N.); [email protected] (R.M.) Abstract: Here, we performed a genome-wide search for methylation sites that contribute to the risk of obesity. We integrated methylation quantitative trait locus (mQTL) data with BMI GWAS information through a SNP-based multiomics approach to identify genomic regions where mQTLs for a methylation site co-localize with obesity risk SNPs. We then tested whether the identified site contributed to BMI through Mendelian randomization. We identified multiple methylation sites causally contributing to the risk of obesity. We validated these findings through a replication stage. By integrating expression quantitative trait locus (eQTL) data, we noted that lower methylation at cg21178254 site upstream of CCNL1 contributes to obesity by increasing the expression of this gene. Higher methylation at cg02814054 increases the risk of obesity by lowering the expression of MAST3, whereas lower methylation at cg06028605 contributes to obesity by decreasing the expression of SLC5A11. Finally, we noted that rare variants within 2p23.3 impact obesity by making the cg01884057 Citation: Nikpay, M.; Ravati, S.; Dent, R.; McPherson, R. -

Defining Phyla: Evolutionary Pathways to Metazoan Body Plans
EVOLUTION & DEVELOPMENT 3:6, 432-442 (2001) Defining phyla: evolutionary pathways to metazoan body plans Allen G. Collins^ and James W. Valentine* Museum of Paleontology and Department of Integrative Biology, University of California, Berkeley, CA 94720, USA 'Author for correspondence (email: [email protected]) 'Present address: Section of Ecology, Befiavior, and Evolution, Division of Biology, University of California, San Diego, La Jolla, CA 92093-0116, USA SUMMARY Phyla are defined by two sets of criteria, one pothesis of Nielsen; the clonal hypothesis of Dewel; the set- morphological and the other historical. Molecular evidence aside cell hypothesis of Davidson et al.; and a benthic hy- permits the grouping of animals into clades and suggests that pothesis suggested by the fossil record. It is concluded that a some groups widely recognized as phyla are paraphyletic, benthic radiation of animals could have supplied the ances- while some may be polyphyletic; the phyletic status of crown tral lineages of all but a few phyla, is consistent with molecu- phyla is tabulated. Four recent evolutionary scenarios for the lar evidence, accords well with fossil evidence, and accounts origins of metazoan phyla and of supraphyletic clades are as- for some of the difficulties in phylogenetic analyses of phyla sessed in the light of a molecular phylogeny: the trochaea hy- based on morphological criteria. INTRODUCTION Molecules have provided an important operational ad- vance to addressing questions about the origins of animal Concepts of animal phyla have changed importantly from phyla. Molecular developmental and comparative genomic their origins in the six Linnaean classis and four Cuvieran evidence offer insights into the genetic bases of body plan embranchements. -

QUANTITATIVE TRAIT LOCUS ANALYSIS: in Several Variants (I.E., Alleles)
TECHNOLOGIES FROM THE FIELD QUANTITATIVE TRAIT LOCUS ANALYSIS: in several variants (i.e., alleles). QTL analysis allows one MULTIPLE CROSS AND HETEROGENEOUS to map, with some precision, the genomic position of STOCK MAPPING these alleles. For many researchers in the alcohol field, the break Robert Hitzemann, Ph.D.; John K. Belknap, Ph.D.; through with respect to the genetic determination of alcohol- and Shannon K. McWeeney, Ph.D. related behaviors occurred when Plomin and colleagues (1991) made the seminal observation that a specific panel of RI mice (i.e., the BXD panel) could be used to identify KEY WORDS: Genetic theory of alcohol and other drug use; genetic the physical location of (i.e., to map) QTLs for behavioral factors; environmental factors; behavioral phenotype; behavioral phenotypes. Because the phenotypes of the different strains trait; quantitative trait gene (QTG); inbred animal strains; in this panel had been determined for many alcohol- recombinant inbred (RI) mouse strains; quantitative traits; related traits, researchers could readily apply the strategy quantitative trait locus (QTL) mapping; multiple cross mapping of RI–QTL mapping (Gora-Maslak et al. 1991). Although (MCM); heterogeneous stock (HS) mapping; microsatellite investigators recognized early on that this panel was not mapping; animal models extensive enough to answer all questions, the emerging data illustrated the rich genetic complexity of alcohol- related phenotypes (Belknap 1992; Plomin and McClearn ntil well into the 1990s, both preclinical and clinical 1993). research focused on finding “the” gene for human The next advance came with the development of diseases, including alcoholism. This focus was rein U microsatellite maps (Dietrich et al.