( 12 ) United States Patent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Atlas Antibodies in Breast Cancer Research Table of Contents
ATLAS ANTIBODIES IN BREAST CANCER RESEARCH TABLE OF CONTENTS The Human Protein Atlas, Triple A Polyclonals and PrecisA Monoclonals (4-5) Clinical markers (6) Antibodies used in breast cancer research (7-13) Antibodies against MammaPrint and other gene expression test proteins (14-16) Antibodies identified in the Human Protein Atlas (17-14) Finding cancer biomarkers, as exemplified by RBM3, granulin and anillin (19-22) Co-Development program (23) Contact (24) Page 2 (24) Page 3 (24) The Human Protein Atlas: a map of the Human Proteome The Human Protein Atlas (HPA) is a The Human Protein Atlas consortium cell types. All the IHC images for Swedish-based program initiated in is mainly funded by the Knut and Alice the normal tissue have undergone 2003 with the aim to map all the human Wallenberg Foundation. pathology-based annotation of proteins in cells, tissues and organs expression levels. using integration of various omics The Human Protein Atlas consists of technologies, including antibody- six separate parts, each focusing on References based imaging, mass spectrometry- a particular aspect of the genome- 1. Sjöstedt E, et al. (2020) An atlas of the based proteomics, transcriptomics wide analysis of the human proteins: protein-coding genes in the human, pig, and and systems biology. mouse brain. Science 367(6482) 2. Thul PJ, et al. (2017) A subcellular map of • The Tissue Atlas shows the the human proteome. Science. 356(6340): All the data in the knowledge resource distribution of proteins across all eaal3321 is open access to allow scientists both major tissues and organs in the 3. -

Amplification of Chromosome 8 Genes in Lung Cancer Onur Baykara1, Burak Bakir1, Nur Buyru1, Kamil Kaynak2, Nejat Dalay3
Journal of Cancer 2015, Vol. 6 270 Ivyspring International Publisher Journal of Cancer 2015; 6(3): 270-275. doi: 10.7150/jca.10638 Research Paper Amplification of Chromosome 8 Genes in Lung Cancer Onur Baykara1, Burak Bakir1, Nur Buyru1, Kamil Kaynak2, Nejat Dalay3 1. Department of Medical Biology, Cerrahpasa Medical Faculty, Istanbul University, Turkey 2. Department of Chest Surgery, Cerrahpasa Medical Faculty, Istanbul University, Turkey 3. Department of Basic Oncology, I.U. Oncology Institute, Istanbul University, Turkey Corresponding author: Prof. Dr. Nejat Dalay, I.U.Oncology Institute, 34093 Capa, Istanbul, Turkey. e-mail : [email protected]; Phone : 90 542 2168861; Fax : 90 212 5348078 © 2015 Ivyspring International Publisher. Reproduction is permitted for personal, noncommercial use, provided that the article is in whole, unmodified, and properly cited. See http://ivyspring.com/terms for terms and conditions. Received: 2014.09.25; Accepted: 2014.12.18; Published: 2015.01.20 Abstract Chromosomal alterations are frequent events in lung carcinogenesis and usually display regions of focal amplification containing several overexpressed oncogenes. Although gains and losses of chromosomal loci have been reported copy number changes of the individual genes have not been analyzed in lung cancer. In this study 22 genes were analyzed by MLPA in tumors and matched normal tissue samples from 82 patients with non-small cell lung cancer. Gene amplifications were observed in 84% of the samples. Chromosome 8 was found to harbor the most frequent copy number alterations. The most frequently amplified genes were ZNF703, PRDM14 and MYC on chromosome 8 and the BIRC5 gene on chromosome 17. The frequency of deletions were much lower and the most frequently deleted gene was ADAM9. -

Logistic Regression Analysis for the Identification of the Metastasis-Associated Signaling Pathways of Osteosarcoma
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 41: 1233-1244, 2018 Logistic regression analysis for the identification of the metastasis-associated signaling pathways of osteosarcoma YANG LIU1, WEI SUN2, XIAOJUN MA2, YUEDONG HAO3, GANG LIU3, XIAOHUI HU3, HOULAI SHANG3, PENGFEI WU3, ZEXUE ZHAO3 and WEIDONG LIU3 1Department of Orthopedics, Affiliated Hospital of Inner Mongolia University for The Nationalities, Tongliao, Inner Mongolia 028007; 2Department of Orthopaedics, Shanghai General Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200080; 3Department of Orthopedics, Huai'an First People's Hospital, Nanjing Medical University, Huai'an, Jiangsu 223300, P.R. China Received February 20, 2017; Accepted September 26, 2017 DOI: 10.3892/ijmm.2018.3360 Abstract. Osteosarcoma (OS) is the most common histological migration were identified as the terms that were significantly type of primary bone cancer. The present study was designed different. ICAM1, PDGFB, PDGFRB and TGFB1 were iden- to identify the key genes and signaling pathways involved in the tified to be enriched in cell migration and regulation of cell metastasis of OS. Microarray data of GSE39055 were down- migration. Nucleotide excision repair and the Wnt signaling loaded from the Gene Expression Omnibus database, which pathway were the metastasis-associated pathways of OS. In included 19 OS biopsy specimens before metastasis (control addition, ICAM1, PDGFB, PDGFRB and TGFB1, which were group) and 18 OS biopsy specimens after metastasis (case involved in cell migration and regulation of cell migration may group). After the differentially expressed genes (DEGs) were affect the metastasis of OS. identified using the Linear Models for Microarray Analysis package, hierarchical clustering analysis and unsupervised Introduction clustering analysis were performed separately, using orange software and the self-organization map method. -

Role of MTDH, FOXM1 and Micrornas in Drug Resistance in Hepatocellular Carcinoma
Diseases 2014, 2, 209-225; doi:10.3390/diseases2030209 OPEN ACCESS diseases ISSN 2079-9721 www.mdpi.com/journal/diseases/ Review Role of MTDH, FOXM1 and microRNAs in Drug Resistance in Hepatocellular Carcinoma Xiangbing Meng 1,2,*, Eric J. Devor 1, Shujie Yang 1, Brandon M. Schickling 3 and Kimberly K. Leslie 1,2 1 Department of Obstetrics and Gynecology, The University of Iowa, Iowa City, IA 52242, USA 2 Holden Comprehensive Cancer Center, The University of Iowa, Iowa City, IA 52242, USA 3 Department of Internal Medicine, The University of Iowa, Iowa City, IA 52242, USA * Author to whom correspondence should be addressed; E-Mail: [email protected]. Received: 16 April 2014; in revised form: 24 June 2014 / Accepted: 25 June 2014 / Published: 1 July 2014 Abstract: Hepatocellular carcinoma (HCC) is one of the most lethal malignancies due to underlying co-morbid cirrhosis and chemo-resistance. Vaccination and improved treatment for hepatitis are the most effective means to reduce the burden of liver cancer worldwide. Expression of biomarkers such as AFP (alpha-fetoprotein), DDK1 (Dickkopf WNT Signaling Pathway Inhibitor 1) and microRNAs in blood are being tested for early screening of liver cancer. Since 2008, sorafenib has been used as the standard molecular targeting agent for HCC. However, overall outcomes for sorafenib alone or in combination with other tyrosine kinase inhibitors are unsatisfactory. Whether simultaneously or sequentially, addiction switches and compensatory pathway activation in HCC, induced by sorafenib treatment, may induce acquired resistance. Forkhead box M1 (FOXM1) and metadherin (MTDH) have been shown to be master regulators of different aspects of tumorigenesis, including angiogenesis, invasion, metastasis and drug resistance. -

Contribution of Alternative Splicing to Breast Cancer Metastasis
Meng et al. J Cancer Metastasis Treat 2019;5:21 Journal of Cancer DOI: 10.20517/2394-4722.2018.96 Metastasis and Treatment Review Open Access Contribution of alternative splicing to breast cancer metastasis Xiangbing Meng1,2, Shujie Yang1,2, Jun Zhang2,3, Huimin Yu1,4 1Department of Obstetrics and Gynecology, University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA. 2Holden Comprehensive Cancer Center, University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA. 3Division of Hematology, Oncology and Blood & Marrow Transplantation, Department of Internal Medicine, University of Iowa Carver College of Medicine, Iowa City, IA 52242, USA. 4Department of Pathogenic Biology, Shenzhen University School of medicine, Shenzhen 518060,China. Correspondence to: Dr. Xiangbing Meng, Department of Obstetrics and Gynecology, The University of Iowa, 375 Newton Road, Iowa City, IA 52242, USA. E-mail: [email protected] How to cite this article: Meng X, Yang S, Zhang J, Yu H. Contribution of alternative splicing to breast cancer metastasis. J Cancer Metastasis Treat 2019;5:21. http://dx.doi.org/10.20517/2394-4722.2018.96 Received: 10 Dec 2018 Accepted: 25 Jan 2019 Published: 22 Mar 2019 Science Editor: William P. Schiemann Copy Editor: Cai-Hong Wang Production Editor: Huan-Liang Wu Abstract Alternative splicing is a major contributor to transcriptome and proteome diversity in eukaryotes. Comparing to normal samples, about 30% more alternative splicing events were recently identified in 32 cancer types included in The Cancer Genome Atlas database. Some alternative splicing isoforms and their encoded proteins contribute to specific cancer hallmarks. In this review, we will discuss recent progress regarding the contributions of alternative splicing to breast cancer metastasis. -

Tumor Suppressive Microrna-375 Regulates Oncogene AEG-1&Sol;MTDH in Head and Neck Squamous Cell Carcinoma (HNSCC)
Journal of Human Genetics (2011) 56, 595–601 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC) Nijiro Nohata1,2, Toyoyuki Hanazawa2, Naoko Kikkawa1,2, Muradil Mutallip1,2, Daiju Sakurai2, Lisa Fujimura3, Kazumori Kawakami4, Takeshi Chiyomaru4, Hirofumi Yoshino4, Hideki Enokida4, Masayuki Nakagawa4, Yoshitaka Okamoto2 and Naohiko Seki1 Our microRNA (miRNA) expression signatures of hypopharyngeal squamous cell carcinoma, maxillary sinus squamous cell carcinoma and esophageal squamous cell carcinoma revealed that miR-375 was significantly reduced in cancer tissues compared with normal epithelium. In this study, we focused on the functional significance of miR-375 in cancer cells and identification of miR-375-regulated novel cancer networks in head and neck squamous cell carcinoma (HNSCC). Restoration of miR-375 showed significant inhibition of cell proliferation and induction of cell apoptosis in SAS and FaDu cell lines, suggesting that miR-375 functions as a tumor suppressor. We adopted genome-wide gene expression analysis to search for miR-375-regulated molecular targets. Gene expression data and luciferase reporter assays revealed that AEG-1/MTDH was directly regulated by miR-375. Cancer cell proliferation was significantly inhibited in HNSCC cells transfected with si-AEG-1/MTDH. In addition, expression levels of AEG-1/MTDH were significantly upregulated in cancer tissues. Therefore, AEG-1/MTDH may function as an oncogene in HNSCC. The identification of novel tumor suppressive miRNA and its regulated cancer pathways could provide new insights into potential molecular mechanisms of HNSCC oncogenesis. -

Engineered Type 1 Regulatory T Cells Designed for Clinical Use Kill Primary
ARTICLE Acute Myeloid Leukemia Engineered type 1 regulatory T cells designed Ferrata Storti Foundation for clinical use kill primary pediatric acute myeloid leukemia cells Brandon Cieniewicz,1* Molly Javier Uyeda,1,2* Ping (Pauline) Chen,1 Ece Canan Sayitoglu,1 Jeffrey Mao-Hwa Liu,1 Grazia Andolfi,3 Katharine Greenthal,1 Alice Bertaina,1,4 Silvia Gregori,3 Rosa Bacchetta,1,4 Norman James Lacayo,1 Alma-Martina Cepika1,4# and Maria Grazia Roncarolo1,2,4# Haematologica 2021 Volume 106(10):2588-2597 1Department of Pediatrics, Division of Stem Cell Transplantation and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 2Stanford Institute for Stem Cell Biology and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 3San Raffaele Telethon Institute for Gene Therapy, Milan, Italy and 4Center for Definitive and Curative Medicine, Stanford School of Medicine, Stanford, CA, USA *BC and MJU contributed equally as co-first authors #AMC and MGR contributed equally as co-senior authors ABSTRACT ype 1 regulatory (Tr1) T cells induced by enforced expression of interleukin-10 (LV-10) are being developed as a novel treatment for Tchemotherapy-resistant myeloid leukemias. In vivo, LV-10 cells do not cause graft-versus-host disease while mediating graft-versus-leukemia effect against adult acute myeloid leukemia (AML). Since pediatric AML (pAML) and adult AML are different on a genetic and epigenetic level, we investigate herein whether LV-10 cells also efficiently kill pAML cells. We show that the majority of primary pAML are killed by LV-10 cells, with different levels of sensitivity to killing. Transcriptionally, pAML sensitive to LV-10 killing expressed a myeloid maturation signature. -

Metabolic Network-Based Stratification of Hepatocellular Carcinoma Reveals Three Distinct Tumor Subtypes
Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes Gholamreza Bidkhoria,b,1, Rui Benfeitasa,1, Martina Klevstigc,d, Cheng Zhanga, Jens Nielsene, Mathias Uhlena, Jan Borenc,d, and Adil Mardinoglua,b,e,2 aScience for Life Laboratory, KTH Royal Institute of Technology, SE-17121 Stockholm, Sweden; bCentre for Host-Microbiome Interactions, Dental Institute, King’s College London, SE1 9RT London, United Kingdom; cDepartment of Molecular and Clinical Medicine, University of Gothenburg, SE-41345 Gothenburg, Sweden; dThe Wallenberg Laboratory, Sahlgrenska University Hospital, SE-41345 Gothenburg, Sweden; and eDepartment of Biology and Biological Engineering, Chalmers University of Technology, SE-41296 Gothenburg, Sweden Edited by Sang Yup Lee, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea, and approved November 1, 2018 (received for review April 27, 2018) Hepatocellular carcinoma (HCC) is one of the most frequent forms of of markers associated with recurrence and poor prognosis (13–15). liver cancer, and effective treatment methods are limited due to Moreover, genome-scale metabolic models (GEMs), collections tumor heterogeneity. There is a great need for comprehensive of biochemical reactions, and associated enzymes and transporters approaches to stratify HCC patients, gain biological insights into have been successfully used to characterize the metabolism of subtypes, and ultimately identify effective therapeutic targets. We HCC, as well as identify drug targets for HCC patients (11, 16–18). stratified HCC patients and characterized each subtype using tran- For instance, HCC tumors have been stratified based on the uti- scriptomics data, genome-scale metabolic networks and network lization of acetate (11). Analysis of HCC metabolism has also led topology/controllability analysis. -

Dysregulation of Micrornas in Breast Cancer and Their Potential Role As
van Schooneveld et al. Breast Cancer Research (2015) 17:21 DOI 10.1186/s13058-015-0526-y REVIEW Open Access Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management Eleni van Schooneveld1*, Hans Wildiers1, Ignace Vergote1, Peter B Vermeulen2, Luc Y Dirix2 and Steven J Van Laere1,2 Abstract MicroRNAs (miRNAs) are an emerging class of gene expression modulators with relevant roles in several biological processes, including cell differentiation, development, apoptosis, and regulation of the cell cycle. Deregulation of those tiny RNA molecules has been described frequently as a major determinant for the initiation and progression of diseases, including cancer. Not only miRNAs but also the enzymes responsible for miRNA processing could be deregulated in cancer. In this review, we address the role of miRNAs in the pathogenesis of breast cancer, since there are oncogenic, tumor-suppressive, and metastatic-influencing miRNAs. Additionally, the different detection platforms and normalization strategies for miRNAs will be discussed. The major part of this review, however, will focus on the capability of miRNAs to act as diagnostic, predictive, or prognostic biomarkers. We will give an overview of their potential to correlate with response to or benefit from a given treatment and we will consider their ability to give information on prognosis in breast cancer. We will focus on miRNAs validated by more than one study or verified in independent cohorts or where results rely on preclinical as well as clinical evidence. As such, we will discuss their potential use in the personalized management of breast cancer. -

Phenethyl Isothiocyanate Suppresses Stemness in the Chemo-And Radio
cancers Article Phenethyl Isothiocyanate Suppresses Stemness in the Chemo- and Radio-Resistant Triple-Negative Breast Cancer Cell Line MDA-MB-231/IR Via Downregulation of Metadherin Yen Thi-Kim Nguyen 1, Jeong Yong Moon 2, Meran Keshawa Ediriweera 2 and Somi Kim Cho 1,2,3,* 1 Interdisciplinary Graduate Program in Advanced Convergence Technology and Science, Jeju National University, Jeju 63243, Korea; [email protected] 2 Subtropical/Tropical Organism Gene Bank, Jeju National University, Jeju 63243, Korea; [email protected] (J.Y.M.); [email protected] (M.K.E.) 3 Faculty of Biotechnology, College of Applied Life Sciences, SARI, Jeju National University, Jeju 63243, Korea * Correspondence: [email protected]; Tel.: +82-010-8660-1842 Received: 6 January 2020; Accepted: 20 January 2020; Published: 22 January 2020 Abstract: Resistance to chemotherapy and radiation therapy is considered a major therapeutic barrier in breast cancer. Cancer stem cells (CSCs) play a prominent role in chemo and radiotherapy resistance. The established chemo and radio-resistant triple-negative breast cancer (TNBC) cell line MDA-MB-231/IR displays greater CSC characteristics than the parental MDA-MB-231 cells. Escalating evidence demonstrates that metadherin (MTDH) is associated with a number of cancer signaling pathways as well as breast cancer therapy resistance, making it an attractive therapeutic target. Kaplan–Meier plot analysis revealed a correlation between higher levels of MTDH and shorter lifetimes in breast cancer and TNBC patients. Moreover, there was a positive correlation between the MTDH and CD44 expression levels in The Cancer Genome Atlas breast cancer database. We demonstrate that MTDH plays a pivotal role in the regulation of stemness in MDA-MB-231/IR cells. -

Efficient and Tumor-Specific Knockdown of MTDH Gene
Yang et al. Breast Cancer Research (2018) 20:113 https://doi.org/10.1186/s13058-018-1042-7 RESEARCHARTICLE Open Access Efficient and tumor-specific knockdown of MTDH gene attenuates paclitaxel resistance of breast cancer cells both in vivo and in vitro Liu Yang1†, Yanhua Tian2†, Wei Sun Leong3†, Heng Song4, Wei Yang1, Meiqi Wang1, Xinle Wang1, Jing Kong3, Baoen Shan1 and Zhengchuan Song1* Abstract Background: Drug resistance of paclitaxel (TAX), the first-line chemotherapy drug for breast cancer, was reported to develop in 90% of patients with breast cancer, especially metastatic breast cancer. Investigating the mechanism of TAX resistance of breast cancer cells and developing the strategy improving its therapeutic efficiency are crucial to breast cancer cure. Methods and Results: We here report an elegant nanoparticle (NP)-based technique that realizes efficient breast cancer treatment of TAX. Using lentiviral vector-mediated gene knockdown, we first demonstrated that TAX therapeutic efficiency was closely correlated with metadherin (MTDH) gene expression in breast cancer cell lines. This finding was also supported by efficacy of TAX treatment in breast cancer patients from our clinical studies. Specifically, TAX treatment became more effective when MTDH expression was decreased in MCF-7 cancer cells by the blocking nuclear factor-kappa B (NF-κB) pathway. Based on these findings, we subsequently synthesized a polymeric NP that could co-deliver MTDH-small interfering RNA (MTDH–siRNA) and TAX into the breast cancer tumors in tumor-bearing mice. The NPs were composed of a cationic copolymer, which wrapped TAX in the inside and adsorbed the negatively charged siRNA on their surface with high drug-loading efficiency and good stability. -
PDF-Document
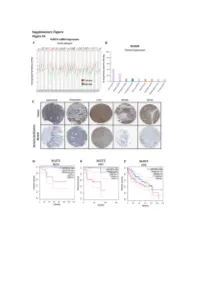
Figure S1: NUDT5 is indicative of poor prognosis and aggressive cancer growth. (A) NUDT5 mRNA expression across full TGCA database, data for tumour (red) and normal (green) tissue from the same tissue origin are shown using TGCA patient datasets with GEPIA interactive webserver [61]. Datasets where NUDT5 mRNA levels are elevated in tumour versus normal as highlighted in red, significantly higher in normal versus tumour are marked in green. Abbreviations of the tumour datasets are given in Table S1. (B) The percentage of positive staining for NUDT5 protein expression in cancer datasets (Human Protein Atlas database [62]). (C) Representative histological staining of NUDT5 in tumour versus normal patient samples from colorectal, pancreatic, liver, breast and renal samples (Human Protein Atlas database [62]). Metadata regarding each of the images are given in Table S2. Kaplan Meyer survival curves stratifying patients based on NUDT5 mRNA expression in (D) Kidney chromophobe (KICH). (E) Adrenalcorticocal carcinoma (ACC). (F) Liver hepatocellular carcinoma (LIHG) (G) Brain lower grade carcinoma (LGG) (H) Kidney renal clear cell carcinoma (KIRC) (I) Kidney renal papillary cell carcinoma (KIRP) were carried out using TGCA database with GEPIA interactive webserver [61]. (J) Recurrence and metastasis analysis in NUDT5 mRNA high expressing patient cohorts from BRCA dataset shown in Figure S1A. (K) Full western blot of NUDT5, PARP1 and FLAG protein levels in NUDT5RES and NUDT5KD cell clones. Figure 2. Analysis of the gene expression changes in 3D compared to 2D culture conditions. (A) PCA analysis of all RNA samples (Table S3) from T47D cells cultured in 2D and 3D conditions. (B) PCA analysis of RNA samples from T47D cells highlighted based on culture conditions.