Characterization of a New Ribosome Associated Quality
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microorganisms – Protists: Euglena
Microorganisms – Protists: Euglena Euglena are unicellular organisms classified into the Kingdom Protista, and the Phylum Euglenophyta. All euglena have chloroplasts and can make their own food by photosynthesis. They are not completely autotrophic though, euglena can also absorb food from their environment. Euglena usually live in quiet ponds or puddles. Euglena move by a flagellum (plural flagella), which is a long whip-like structure that acts like a little motor. The flagellum is located on the anterior (front) end, and twirls in such a way as to pull the cell through the water. It is attached at an inward pocket called the reservoir. Color and label the reservoir grey. Color and label the flagellum black. The Euglena is unique in that it is both heterotrophic (must consume food) and autotrophic (can make its own food). Chloroplasts within the euglena trap sunlight that is used for photosynthesis and can be seen as several rod-like structures throughout the cell. Color and label the chloroplasts green. Euglena also have an eyespot at the anterior end that detects light, it can be seen near the reservoir. This helps the euglena find bright areas to gather sunlight to make their food. Color and label the eyespot red. Euglena can also gain nutrients by absorbing them across their cell membrane, hence they become heterotrophic when light is not available, and they cannot photosynthesize. The euglena has a stiff pellicle outside the cell membrane that helps it keep its shape, though the pellicle is somewhat flexible, and some euglena can be observed scrunching up and moving in an inchworm type fashion. -

A Physicochemical Perspective of Aging from Single-Cell Analysis Of
TOOLS AND RESOURCES A physicochemical perspective of aging from single-cell analysis of pH, macromolecular and organellar crowding in yeast Sara N Mouton1, David J Thaller2, Matthew M Crane3, Irina L Rempel1, Owen T Terpstra1, Anton Steen1, Matt Kaeberlein3, C Patrick Lusk2, Arnold J Boersma4*, Liesbeth M Veenhoff1* 1European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, Groningen, Netherlands; 2Department of Cell Biology, Yale School of Medicine, New Haven, United States; 3Department of Pathology, School of Medicine, University of Washington, Seattle, United States; 4DWI-Leibniz Institute for Interactive Materials, Aachen, Germany Abstract Cellular aging is a multifactorial process that is characterized by a decline in homeostatic capacity, best described at the molecular level. Physicochemical properties such as pH and macromolecular crowding are essential to all molecular processes in cells and require maintenance. Whether a drift in physicochemical properties contributes to the overall decline of homeostasis in aging is not known. Here, we show that the cytosol of yeast cells acidifies modestly in early aging and sharply after senescence. Using a macromolecular crowding sensor optimized for long-term FRET measurements, we show that crowding is rather stable and that the stability of crowding is a stronger predictor for lifespan than the absolute crowding levels. Additionally, in aged cells, we observe drastic changes in organellar volume, leading to crowding on the *For correspondence: micrometer scale, which we term organellar crowding. Our measurements provide an initial [email protected] framework of physicochemical parameters of replicatively aged yeast cells. (AJB); [email protected] (LMV) Competing interest: See Introduction page 19 Cellular aging is a process of progressive decline in homeostatic capacity (Gems and Partridge, Funding: See page 19 2013; Kirkwood, 2005). -

A Signal-Anchor Sequence Stimulates Signal Recognition Particle Binding to Ribosomes from Inside the Exit Tunnel
A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel Uta Berndta,b,1, Stefan Oellerera,b,c,1, Ying Zhanga,b,c, Arthur E. Johnsond, and Sabine Rosperta,b,2 aInstitute of Biochemistry and Molecular Biology and bCenter for Biological Signalling Studies, University of Freiburg, Stefan-Meier-Strasse 17, D-79104 Freiburg, Germany; cFakulta¨t fu¨ r Biologie, University of Freiburg, Scha¨nzlestrasse 1, D-79104 Freiburg, Germany; and dDepartment of Molecular and Cellular Medicine, Texas A&M Health Science Center, 116 Reynolds Medical Building, College Station, TX 77843 Edited by Arthur Horwich, Yale University School of Medicine, New Haven, CT, and approved December 15, 2008 (received for review August 29, 2008) Sorting of eukaryotic membrane and secretory proteins depends direct interaction between SRP and the nascent chain. Previous on recognition of ribosome-bound nascent chain signal sequences studies have addressed the question of whether or not specific by the signal recognition particle (SRP). The current model suggests amino acid sequences of segments inside the tunnel can further that the SRP cycle is initiated when a signal sequence emerges from the affinity of SRP for RNCs (9, 10). Because signal sequences the ribosomal tunnel and binds to SRP. Then elongation is slowed would be prime candidates for such effects, this possibility was until the SRP-bound ribosome–nascent chain complex (RNC) is tested in the eukaryotic system by using RNCs carrying prep- targeted to the SRP receptor in the endoplasmic reticulum (ER) rolactin, a secreted protein with a cleavable signal sequence. membrane. The RNC is then transferred to the translocon, SRP is However, when nascent preprolactin was too short to exit the released, and translation resumes. -

The Ins and Outs of Autophagic Ribosome Turnover
Biochemistry, Biophysics and Molecular Biology Publications Biochemistry, Biophysics and Molecular Biology 2019 The Ins and Outs of Autophagic Ribosome Turnover Zakayo Kazibwe Iowa State University, [email protected] Ang-Yu Liu Iowa State University, [email protected] Gustavo C. Macintosh Iowa State University, [email protected] Diane C. Bassham Iowa State University, [email protected] Follow this and additional works at: https://lib.dr.iastate.edu/bbmb_ag_pubs Part of the Cell and Developmental Biology Commons, Genetics Commons, and the Molecular Biology Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ bbmb_ag_pubs/260. For information on how to cite this item, please visit http://lib.dr.iastate.edu/howtocite.html. This Article is brought to you for free and open access by the Biochemistry, Biophysics and Molecular Biology at Iowa State University Digital Repository. It has been accepted for inclusion in Biochemistry, Biophysics and Molecular Biology Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. The Ins and Outs of Autophagic Ribosome Turnover Abstract Ribosomes are essential for protein synthesis in all organisms and their biogenesis and number are tightly controlled to maintain homeostasis in changing environmental conditions. While ribosome assembly and quality control mechanisms have been extensively studied, our understanding of ribosome degradation is limited. In yeast or animal cells, ribosomes are degraded after transfer into the vacuole or lysosome by ribophagy or nonselective autophagy, and ribosomal RNA can also be transferred directly across the lysosomal membrane by RNautophagy. In plants, ribosomal RNA is degraded by the vacuolar T2 ribonuclease RNS2 after transport by autophagy-related mechanisms, although it is unknown if a selective ribophagy pathway exists in plants. -

Cooperation of Mitochondrial and ER Factors in Quality Control of Tail
RESEARCH ARTICLE Cooperation of mitochondrial and ER factors in quality control of tail-anchored proteins Verena Dederer1, Anton Khmelinskii1,2, Anna Gesine Huhn1, Voytek Okreglak3, Michael Knop1,4, Marius K Lemberg1* 1Centre for Molecular Biology of Heidelberg University (ZMBH), DKFZ-ZMBH Alliance, Heidelberg, Germany; 2Institute of Molecular Biology (IMB), Mainz, Germany; 3Calico Life Sciences LLC, South San Francisco, United States; 4Cell Morphogenesis and Signal Transduction, German Cancer Research Center (DKFZ), Heidelberg, Germany Abstract Tail-anchored (TA) proteins insert post-translationally into the endoplasmic reticulum (ER), the outer mitochondrial membrane (OMM) and peroxisomes. Whereas the GET pathway controls ER-targeting, no dedicated factors are known for OMM insertion, posing the question of how accuracy is achieved. The mitochondrial AAA-ATPase Msp1 removes mislocalized TA proteins from the OMM, but it is unclear, how Msp1 clients are targeted for degradation. Here we screened for factors involved in degradation of TA proteins mislocalized to mitochondria. We show that the ER-associated degradation (ERAD) E3 ubiquitin ligase Doa10 controls cytoplasmic level of Msp1 clients. Furthermore, we identified the uncharacterized OMM protein Fmp32 and the ectopically expressed subunit of the ER-mitochondria encounter structure (ERMES) complex Gem1 as native clients for Msp1 and Doa10. We propose that productive localization of TA proteins to the OMM is ensured by complex assembly, while orphan subunits are extracted by Msp1 and eventually degraded by Doa10. *For correspondence: DOI: https://doi.org/10.7554/eLife.45506.001 [email protected]. de Competing interests: The authors declare that no Introduction competing interests exist. Correct localization of proteins is essential to ensure their functionality and to establish the identity Funding: See page 19 of individual cellular organelles. -

CILIA of a DISTINCTIVE STRUCTURE (G + O) in ENDOCRINE and OTHER TISSUES
Postgrad Med J: first published as 10.1136/pgmj.42.489.403 on 1 July 1966. Downloaded from POSTGRAD MED. J. (1966), 42, 403 CILIA OF A DISTINCTIVE STRUCTURE (g + o) IN ENDOCRINE AND OTHER TISSUES A. R. CURRIE, BSC., F.R.C.P. (Edin. and Glas.), F.C.Path., F.R.S.E. D. N. WHEATLEY, B.Sc., Ph.D., 'MI.Biol. Department of Pathology, University Medical Buildings, Foresterhill, Aberdeen. CILIA or flagella control and effect the move- ear (Gray, 1960), and in sense cells of Pecten ment of many unicellular organisms, for (Miller, 1958) and crab (Whitear, 1960). They example Paramecium; they may also cause have been more frequently reported, however, in movement of the medium surrounding cells vertebrate tissues-particularly mammalian- as in flame cells of Platyhelminuthes and in the but this may be due to the fact that these tissues epidielium of the respiratory tract and fallopian have been more intensively studied with the tu)bes in tthe human subject. The fine structure electron microscope (Talble 1). of these cilia is well known and is essentially the same from the 'lowest ciliated creature to the highest mammals (for a brief review see Structure of 9 + 0 cilia 1962). Grimstone, 9 + 0 cilia in mammalian tissues differ from The cilia -are characterized by a 9 + 2 fibre the 9 + 2 form in that they are associated with copyright. patern (Fig. '1) and a single basal body; the a 'diplosome that is a pasir of centrioles, one basal 'body, which has been referred to variously usually lying at right angles to the other. -

Membrane Structure and Function
Chapter 7 Membrane Structure and Function Lecture Outline Overview: Life at the Edge • The plasma membrane separates the living cell from its surroundings. • This thin barrier, 8 nm thick, controls traffic into and out of the cell. • Like all biological membranes, the plasma membrane is selectively permeable, allowing some substances to cross more easily than others. • The formation of a membrane that encloses a solution different from the surrounding solution while still permitting the uptake of nutrients and the elimination of waste products was a key event in the evolution of life. • The ability of the cell to discriminate in its chemical exchanges with its environment is fundamental to life. • It is the plasma membrane and its component molecules that make this selectivity possible. Concept 7.1 Cellular membranes are fluid mosaics of lipids and proteins. • The main macromolecules in membranes are lipids and proteins, but carbohydrates are also important. • The most abundant lipids are phospholipids. • Phospholipids and most other membrane constituents are amphipathic molecules, which have both hydrophobic and hydrophilic regions. Membrane models have evolved to fit new data. • The arrangement of phospholipids and proteins in biological membranes is described by the fluid mosaic model. • In this model, the membrane is a fluid structure with a “mosaic” of various proteins embedded in or attached to a double layer (bilayer) of phospholipids. • Models of membranes were developed long before membranes were first seen with electron microscopes in the 1950s. • In 1915, membranes isolated from red blood cells were chemically analyzed and found to be composed of lipids and proteins. • In 1925, E. -

Congratulations!!! You Have Found the Vacuole!
CONGRATULATIONS!!! YOU HAVE FOUND THE VACUOLE! How is the book room like the vacuole? What would represent the food, the water, and the waste? The vacuole is a very important cell organelle to all types of cells but it has the most important job in the plant cell. You can think of the vacuole as sort of a storage container, it can store water, food, and waste. Vacuoles also isolate unwanted bacteria and dispose of them so they do not harm the cell. The vacuoles in both plant and animal cells are made from Golgi bodies inside the cell. Vacuoles are made up of multiple membrane vesicles. Certain substances created by the cell that are unwanted and could be harmful to other organelles are isolated and taken care of by being sent to the Golgi bodies. Vacuoles bring their stored material to any organelle inside the cell that needs it or to other cells if they need the stored material. Vacuoles in plant and animal cells are different in size and numbers but they ultimately have the same functions. In a plant cell, there is one central vacuole that is very big and has lots of room. Animal cells have many vacuoles but they are really small. A plant cells vacuole would process chlorophyll and chloroplast while the animal cell does not even have chloroplast organelles. It is estimated that the central vacuole in a plant cell takes up 80% of the cell. Something to help you remember that it stores water is to think of a camel hump. When the camel has humps that means that it has water. -

Content and Vacuole/Extravacuole Distribution of Neutral Sugars, Free
Plant Physiol. (1979) 64, 88-93 0032-0889/79/64/0088/06/$00.50/0 Content and Vacuole/Extravacuole Distribution of Neutral Sugars, Free Amino Acids, and Anthocyanin in Protoplasts1 Received for publication December 26, 1978 and in revised form March 15, 1979 GEORGE J. WAGNER Department ofBiology, Brookhaven National Laboratory, Upton, New York 11973 ABSTRACT Hybrid (petals from anthesis stage flowers) and Tulipa cv. Red Shine for amino acid and other studies, and cv. Most Miles for Neutral sugar, free amino acid, and anthocyanin levels and vacuole/ sugar analysis (petals from 1- to 2-day postanthesis flowers and extravacuole distribution were determined for Hippeastum and Tulipa oldest leaves from 5-day postanthesis plants). Tulip bulbs were petal and Tudipa leaf protoplasts. Glucose and fructose, the predominant obtained from K. Van Bourgardein and Sons, Inc., Babylon, N.Y., neutral monosaccharides observed, were primarily vacuolar in location. and Hippeastrum from Jackson & Perkins Co. Tulip bulbs were Glutamine, the predominant free amino acid found, was primarily extra- rooted at 9 C then held at 0.6 C as long as 10 months before vacuolar. y-Methyleneglutamate was identified as a major constituent of induction of vegetative and floral growth at 22 C. In this way Tudpa protoplasts. Qualitative characterization of Hippeastrum petal and flowers and leaves were made available for 10 months of the year. vacuole organic acids indicated the presence of oxalic, malic, citric, and Hippeastrum flowers were made available year round. For this isocitric acids. Data are presented which indicate that vacuoles obtained plant, postflowering vegetative growth (3 months) was terminated by gentle osmotic shock of protoplasts in dibasic phosphate have good by drying the bulbs followed by vernalization for a minimum of purity and retain their contents. -
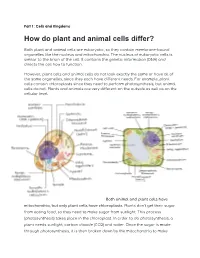
How Do Plant and Animal Cells Differ?
Part 1: Cells and Kingdoms How do plant and animal cells differ? Both plant and animal cells are eukaryotic, so they contain membrane-bound organelles like the nucleus and mitochondria. The nucleus of eukaryotic cells is similar to the brain of the cell. It contains the genetic information (DNA) and directs the cell how to function. However, plant cells and animal cells do not look exactly the same or have all of the same organelles, since they each have different needs. For example, plant cells contain chloroplasts since they need to perform photosynthesis, but animal cells do not. Plants and animals are very different on the outside as well as on the cellular level. Both animal and plant cells have mitochondria, but only plant cells have chloroplasts. Plants don’t get their sugar from eating food, so they need to make sugar from sunlight. This process (photosynthesis) takes place in the chloroplast. In order to do photosynthesis, a plant needs sunlight, carbon dioxide (CO2) and water. Once the sugar is made through photosynthesis, it is then broken down by the mitochondria to make energy for the cell. Because animals get sugar from the food they eat, they do not need chloroplasts: just mitochondria. Both plant and animal cells have vacuoles. A plant cell contains a large, singular vacuole that is used for storage of water and nutrients. It also helps maintain the shape of the cell. In contrast, animal cells have many, smaller vacuoles, which also are used for storage of water and nutrients. Plant cells have a cell wall, as well as a cell membrane. -

Cellular Structure and Function Section ●3 Structures and Organelles
chapter 7 Cellular Structure and Function section ●3 Structures and Organelles Before You Read -!). )DEA The eukaryotic cell contains For cells to function correctly, each part must do its job. organelles. Members of families have jobs or chores that help the whole What You’ll Learn family. On the lines below, list your family members and their differences in the structures of jobs. plant and animal cells Read to Learn Identify the Parts Highlight Cytoplasm and Cytoskeleton each cell structure as you read The environment inside the plasma membrane is a about it. Underline the function semifl uid material called cytoplasm. Scientists once thought of each part. the organelles of eukaryotic cells fl oated freely in the cell’s cytoplasm. As technology improved, scientists discovered more about cell structures. They discovered a structure within the cytoplasm called the cytoskeleton. The cytoskeleton is a network of long, thin protein fi bers that provide an anchor for organelles inside the cell. The cell’s shape and movement depend on the cytoskeleton. Two types of protein fi bers make up the cytoskeleton. Microtubules are long, hollow protein cylinders that form a fi rm skeleton for the cell. They assist in moving substances within the cell. Microfi laments are thin protein threads that help give the cell shape and enable the entire cell or parts of the cell to move. 1. Name one cell function that takes place in organelles. Cell Structures Copyright © Glencoe/McGraw-Hill, a division of The McGraw-Hill Companies, Inc. Companies, a division of The McGraw-Hill © Glencoe/McGraw-Hill, Copyright All chemical processes of a typical eukaryotic cell take place in the organelles, which move around in the cell’s cytoplasm. -

I V Molecular Mechanisms of Notochord Vacuole Formation And
Molecular Mechanisms of Notochord Vacuole Formation and Their Role in Zebrafish Development by Kathryn Leigh Ellis Department of Cell Biology Duke University Date:_______________________ Approved: ___________________________ Michel Bagnat, Advisor ___________________________ Kenneth Poss, Chair ___________________________ Christopher Nicchitta ___________________________ Jun Chen ___________________________ Raphael Valdivia Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Cell Biology in the Graduate School of Duke University 2014 i v ABSTRACT Molecular Mechanisms of Notochord Vacuole Formation and Their Role in Zebrafish Development by Kathryn Leigh Ellis Department of Cell Biology Duke University Date:_______________________ Approved: ___________________________ Michel Bagnat, Advisor ___________________________ Kenneth Poss, Chair ___________________________ Christopher Nicchitta ___________________________ Jun Chen ___________________________ Raphael Valdivia An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Cell Biology in the Graduate School of Duke University 2014 Copyright by Kathryn Leigh Ellis 2014 Abstract The notochord plays critical structural and signaling roles during vertebrate development. At the center of the vertebrate notochord is a large fluid- filled organelle, the notochord vacuole. While these highly conserved intracellular structures have been described for decades, little is known about the molecular mechanisms involved in their biogenesis and maintenance. Here we show that zebrafish notochord vacuoles are specialized lysosome-related organelles whose formation and maintenance requires late endosomal trafficking regulated by the vacuole-specific Rab32a, and H+-ATPase-dependent acidification. We establish that notochord vacuoles are required for body axis elongation during embryonic development and identify a novel role for notochord vacuoles in spine morphogenesis.