Expression of Transcript Variants of PTGS1 and PTGS2 Genes Among Patients with Chronic Rhinosinusitis with Nasal Polyps
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diclofenac Enhances Docosahexaenoic Acid-Induced Apoptosis in Vitro in Lung Cancer Cells
cancers Article Diclofenac Enhances Docosahexaenoic Acid-Induced Apoptosis in Vitro in Lung Cancer Cells Rosemary A. Poku 1, Kylee J. Jones 2, Megan Van Baren 2 , Jamie K. Alan 3 and Felix Amissah 2,* 1 Department of Foundational Sciences, College of Medicine, Central Michigan University, Warriner Hall, 319, Mt Pleasant, MI 48859, USA; [email protected] 2 Department of Pharmaceutical Sciences, Ferris State University, College of Pharmacy, 220 Ferris Dr, Big Rapids, MI 49307, USA; [email protected] (K.J.J.); [email protected] (M.V.B.) 3 Department of Pharmacology and Toxicology, Michigan State University, East Lansing, MI 48824, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-231-591-3790 Received: 9 September 2020; Accepted: 17 September 2020; Published: 20 September 2020 Simple Summary: Polyunsaturated fatty acids (PUFAs) and non-steroidal anti-inflammatory drugs (NSAIDs) have limited anticancer capacities when used alone. We examined whether combining NSAIDs with docosahexaenoic (DHA) would increase their anticancer activity on lung cancer cell lines. Our results indicate that combining DHA and NSAIDs increased their anticancer activities by altering the expression of critical proteins in the RAS/MEK/ERK and PI3K/Akt pathways. The data suggest that DHA combined with low dose diclofenac provides more significant anticancer potential, which can be further developed for chemoprevention and adjunct therapy in lung cancer. Abstract: Polyunsaturated fatty acids (PUFAs) and non-steroidal anti-inflammatory drugs (NSAIDs) show anticancer activities through diverse molecular mechanisms. However, the anticancer capacities of either PUFAs or NSAIDs alone is limited. We examined whether combining NSAIDs with docosahexaenoic (DHA), commonly derived from fish oils, would possibly synergize their anticancer activity. -

AA Metabolism, 139 A2b1, 69, 70, 318–323 ABCB1 Gene, 176 Abciximab
Index A aIIbb3, 60, 62, 69, 70, 73, 74, 79, 80 AA. See Arachidonic acid (AA) AJW202, 293 AA metabolism, 139 Akt-1, 345 a2b1, 69, 70, 318–323 Akt-2, 345 ABCB1 gene, 176 ALX-0081, 296 Abciximab, 206–208, 497, 500, 501, 504–505, ALX-0681, 298 509, 513, 514, 525 Anagrelide, 227, 229 approval, 206 Ankle brachial indexes (ABIs), 549, 587, EPIC trial, 207 595–597 EPILOG study, 207 framingham risk score, 551 EPISTENT study, 207 screening, 550 platelet binding, 207 sensitivity, 551 thrombocytopenia, 206 specificity, 551 in unstable angina, 208 vascular risk, 550 ABIs. See Ankle brachial indexes (ABIs) Antagomirs, 439 Acetylsalicylic acid (aspirin), 137–158 Antibodies, 317 ACS. See Acute coronary syndromes (ACS) Anticoagulants, 525, 537 Acute coronary syndromes (ACS), 288 Anticoagulation, 533, 534 Acute ischemic stroke, 143, 149–150 Antioxidant, 233 Acute myocardial infarction, 142, 143, 149, Antiplatelet agents, 263, 553 153, 157 Antithrombotic Trialist’s ADAMTS-13, 93 Collaboration, 553 Adenosine diphosphate (ADP), 37, 88, 96, aspirin, 553 166, 473 clopidogrel, 553 Adenosine triphosphate (ATP), 37 dipyridamole, 553 Adhesion, 90, 112–118, 125 meta-analysis, 560 Adhesion molecules infiltration, 264 picotamide, 553 Adiponectin, 315 risk reduction, 553 ADP. See Adenosine diphosphate (ADP) ticlopidine, 553 ADP inhibitors (or P2Y12 blockers), 497–499, vascular events, 553 501–505, 507–509, 513, 514 Antiplatelet therapy, 472 ADP-receptor antagonists, 169 Apixaban, 535, 539 Adverse effects, 153–156 Aptamers, 299–301 Aegyptin, 327 Arachidonic acid (AA), 474 P. Gresele et al. (eds.), Antiplatelet Agents, Handbook of Experimental 607 Pharmacology 210, DOI 10.1007/978-3-642-29423-5, # Springer-Verlag Berlin Heidelberg 2012 608 Index ARC1779, 299–300 TXS signaling, 279 ARC15105, 300 Cardiovascular death, 248 AR-C69931MX, 456 Carotid endarterectomy (ACE) inhibition, Aspirin, 474, 496, 498–499, 501–502, 143, 156 504–507, 511–514, 520, 522, Carotid stenosis, 523 527, 531, 533–537, 539, CCBs. -

OBE022, an Oral and Selective Prostaglandin F2α Receptor Antagonist As an Effective and Safe Modality for the Treatment of Pret
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/05/18/jpet.118.247668.DC1 1521-0103/366/2/349–364$35.00 https://doi.org/10.1124/jpet.118.247668 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:349–364, August 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics OBE022, an Oral and Selective Prostaglandin F2a Receptor Antagonist as an Effective and Safe Modality for the Treatment of Preterm Labor s Oliver Pohl, André Chollet, Sung Hye Kim, Lucia Riaposova, François Spézia, Frédéric Gervais, Philippe Guillaume, Philippe Lluel, Murielle Méen, Frédérique Lemaux, Vasso Terzidou, Phillip R. Bennett, and Jean-Pierre Gotteland ObsEva SA, Plan-les-Ouates, Geneva, Switzerland (O.P., A.C., J.-P.G.); Imperial College London, Parturition Research Group, Institute of Reproductive and Developmental Biology,HammersmithHospitalCampus,EastActon,London,UnitedKingdom(S.H.K.,L.R., V.T., P.R.B.); Citoxlab, Evreux, France (F.S., F.G.); Porsolt Research Laboratory, Le Genest-Saint-Isle, France (P.G.); Urosphere SAS, Toulouse, France (P.L., M.M.); BioTrial, Rennes, France (F.L.); and André Chollet Consulting, Tannay, Switzerland (A.C.) Downloaded from Received February 26, 2018; accepted May 15, 2018 ABSTRACT Preterm birth is the major challenge in obstetrics, affecting aggregation. In in vitro studies, OBE002 inhibited sponta- ∼ 10% of pregnancies. Pan-prostaglandin synthesis inhibitors neous, oxytocin- and PGF2a-induced human myometrial jpet.aspetjournals.org [nonsteroidal anti-inflammatory drugs (NSAIDs)] prevent preterm contractions alone and was more effective in combination labor and prolong pregnancy but raise concerns about fetal renal with atosiban or nifedipine. -

Inhibition of PTGS1 Promotes Osteogenic Differentiation Of
Wang et al. Stem Cell Research & Therapy (2019) 10:57 https://doi.org/10.1186/s13287-019-1167-3 RESEARCH Open Access Inhibition of PTGS1 promotes osteogenic differentiation of adipose-derived stem cells by suppressing NF-kB signaling Yuejun Wang1,2, Yunsong Liu1,2, Min Zhang1,2, Longwei Lv1,2, Xiao Zhang1,2, Ping Zhang1,2* and Yongsheng Zhou1,2* Abstract Background: Tissue inflammation is an important problem in the field of human adipose-derived stem cell (ASC)- based therapeutic bone regeneration. Many studies indicate that inflammatory cytokines are disadvantageous for osteogenic differentiation and bone formation. Therefore, overcoming inflammation would be greatly beneficial in promoting ASC-mediated bone regeneration. The present study aims to investigate the potential anti-inflammatory role of Prostaglandin G/H synthase 1 (PTGS1) during the osteogenic differentiation of ASCs. Methods: We performed TNFα treatment to investigate the response of PTGS1 to inflammation. Loss- and gain-of- function experiments were applied to investigate the function of PTGS1 in the osteogenic differentiation of ASCs ex vivo and in vivo. Western blot and confocal analyses were used to determine the molecular mechanism of PTGS1-regulated osteogenic differentiation. Results: Our work demonstrates that PTGS1 expression is significantly increased upon inflammatory cytokine treatment. Both ex vivo and in vivo studies indicate that PTGS1 is required for the osteogenic differentiation of ASCs. Mechanistically, we show that PTGS1 regulates osteogenesis of ASCs via modulating the NF-κB signaling pathway. Conclusions: Collectively, this work confirms that the PTGS1-NF-κB signaling pathway is a novel molecular target for ASC-mediated regenerative medicine. Keywords: PTGS1, NF-κB, Osteogenic differentiation, ASCs Background tissue. -

Role of Arachidonic Acid and Its Metabolites in the Biological and Clinical Manifestations of Idiopathic Nephrotic Syndrome
International Journal of Molecular Sciences Review Role of Arachidonic Acid and Its Metabolites in the Biological and Clinical Manifestations of Idiopathic Nephrotic Syndrome Stefano Turolo 1,* , Alberto Edefonti 1 , Alessandra Mazzocchi 2, Marie Louise Syren 2, William Morello 1, Carlo Agostoni 2,3 and Giovanni Montini 1,2 1 Fondazione IRCCS Ca’ Granda-Ospedale Maggiore Policlinico, Pediatric Nephrology, Dialysis and Transplant Unit, Via della Commenda 9, 20122 Milan, Italy; [email protected] (A.E.); [email protected] (W.M.); [email protected] (G.M.) 2 Department of Clinical Sciences and Community Health, University of Milan, 20122 Milan, Italy; [email protected] (A.M.); [email protected] (M.L.S.); [email protected] (C.A.) 3 Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Pediatric Intermediate Care Unit, 20122 Milan, Italy * Correspondence: [email protected] Abstract: Studies concerning the role of arachidonic acid (AA) and its metabolites in kidney disease are scarce, and this applies in particular to idiopathic nephrotic syndrome (INS). INS is one of the most frequent glomerular diseases in childhood; it is characterized by T-lymphocyte dysfunction, alterations of pro- and anti-coagulant factor levels, and increased platelet count and aggregation, leading to thrombophilia. AA and its metabolites are involved in several biological processes. Herein, Citation: Turolo, S.; Edefonti, A.; we describe the main fields where they may play a significant role, particularly as it pertains to their Mazzocchi, A.; Syren, M.L.; effects on the kidney and the mechanisms underlying INS. AA and its metabolites influence cell Morello, W.; Agostoni, C.; Montini, G. -

Success of Prostaglandin E2 in Structure–Function Is a Challenge for Structure-Based Therapeutics
COMMENTARY Success of prostaglandin E2 in structure–function is a challenge for structure-based therapeutics Charles N. Serhan*† and Bruce Levy*‡ *Center for Experimental Therapeutics and Reperfusion Injury, Department of Anesthesiology, Perioperative and Pain Medicine and ‡Critical Care and Pulmonary Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115 rostaglandin (PG) E2 is almost ubiquitous in humans and evokes potent diverse actions. Utility is the price of its perfec- Ption. PGE is a founding member of the 2 PGs, a class of mediators that belongs to the still growing family of bioactive au- tacoids known as the eicosanoids (1–3). The main classes include enzymatically generated products such as thrombox- anes, leukotrienes, lipoxins, and EETs, as well as others that are produced via nonenzymatic mechanisms, e.g., isopros- tanes and cyclopentaeone PGs that are increasing in number and appreciation (4, 5). PGE2 regulates key responses in the major human systems including re- productive, gastrointestinal, neuroendo- crine, and immune (Fig. 1). Formed by conversion of arachidonic acid via cyclo- oxygenase (COX) and specific synthases, Fig. 1. Diverse actions of PGE2 and selective targeted biosynthesis in inflammation. (Inset) Eicosanoid family major enzymatic classes of cyclooxygenases and lipoxygenase pathways (see text for details). PGE2 stereospecifically exerts potent (nano- to micromolar range) tissue- and cell type-selective actions (1–6). The importance of PGs in inflammation was from protecting gastrointestinal mucosa electrophoresis that this lipid-soluble brought into view by the discovery of J. to regulating smooth muscle and fever, activity behaved as an acid. Vane and colleagues (7) that nonsteroi- set a steep challenge for designer drug Bergstro¨m’s main research was in bile dal antiinflammatory drugs (NSAIDs) hunters to achieve, namely selectivity acids and steroids. -

Downloaded from Bioscientifica.Com at 09/25/2021 12:14:58PM Via Free Access
ID: 19-0149 8 6 Q Pang et al. PHO patients with soft tissue 8:6 736–744 giant tumor caused by HPGD mutation RESEARCH The first case of primary hypertrophic osteoarthropathy with soft tissue giant tumors caused by HPGD loss-of-function mutation Qianqian Pang1,2, Yuping Xu1,3, Xuan Qi1, Yan Jiang1, Ou Wang1, Mei Li1, Xiaoping Xing1, Ling Qin2 and Weibo Xia1 1Department of Endocrinology, Key Laboratory of Endocrinology, Ministry of Health, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Beijing, China 2Musculoskeletal Research Laboratory and Bone Quality and Health Assessment Centre, Department of Orthopedics & Traumatology, The Chinese University of Hong Kong, Hong Kong SAR, Hong Kong 3Department of Endocrinology, The First Affiliated Hospital of Shanxi Medicalniversity, U Taiyuan, Shanxi, China Correspondence should be addressed to L Qin or W Xia: [email protected] or [email protected] Abstract Background: Primary hypertrophic osteoarthropathy (PHO) is a rare genetic multi-organic Key Words disease characterized by digital clubbing, periostosis and pachydermia. Two genes, f primary hypertrophic HPGD and SLCO2A1, which encodes 15-hydroxyprostaglandin dehydrogenase (15-PGDH) osteoarthropathy and prostaglandin transporter (PGT), respectively, have been reported to be related to f PHO PHO. Deficiency of aforementioned two genes leads to failure of prostaglandin E2 (PGE2) f HPGD mutation degradation and thereby elevated levels of PGE2. PGE2 plays an important role in f soft tumor tumorigenesis. Studies revealed a tumor suppressor activity of 15-PGDH in tumors, such f COX2 selective inhibitor treatment as lung, bladder and breast cancers. However, to date, no HPGD-mutated PHO patients presenting concomitant tumor has been documented. -

S42003-021-02488-1.Pdf
ARTICLE https://doi.org/10.1038/s42003-021-02488-1 OPEN In situ imaging reveals disparity between prostaglandin localization and abundance of prostaglandin synthases ✉ Kyle D. Duncan 1,5, Xiaofei Sun 2,3,5, Erin S. Baker 4, Sudhansu K. Dey 2,3 & Ingela Lanekoff 1 Prostaglandins are important lipids involved in mediating many physiological processes, such as allergic responses, inflammation, and pregnancy. However, technical limitations of in-situ prostaglandin detection in tissue have led researchers to infer prostaglandin tissue dis- tributions from localization of regulatory synthases, such as COX1 and COX2. Herein, we apply a novel mass spectrometry imaging method for direct in situ tissue localization of 1234567890():,; prostaglandins, and combine it with techniques for protein expression and RNA localization. We report that prostaglandin D2, its precursors, and downstream synthases co-localize with the highest expression of COX1, and not COX2. Further, we study tissue with a conditional deletion of transformation-related protein 53 where pregnancy success is low and confirm that PG levels are altered, although localization is conserved. Our studies reveal that the abundance of COX and prostaglandin D2 synthases in cellular regions does not mirror the regional abundance of prostaglandins. Thus, we deduce that prostaglandins tissue localization and abundance may not be inferred by COX or prostaglandin synthases in uterine tissue, and must be resolved by an in situ prostaglandin imaging. 1 Department of Chemistry-BMC, Uppsala University, Uppsala, Sweden. 2 Division of Reproductive Sciences, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229, USA. 3 College of Medicine, University of Cincinnati, Cincinnati, OH 45221, USA. -

Cyclooxygenase-Independent Inhibition by Aspirin
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 11091-11096, October 1996 Medical Sciences Leukocyte lipid body formation and eicosanoid generation: Cyclooxygenase-independent inhibition by aspirin (nonsteroidal antiinflammatory agents/cyclooxygenase knockout/leukotrienes/prostaglandin endoperoxide H synthase/lipoxygenase) PATRICIA T. BOZZA*, JENNIFER L. PAYNE*, ScoTr G. MORHAMt, ROBERT LANGENBACHt, OLIVER SMITHIESt, AND PETER F. WELLER*§ *Harvard Thorndike Laboratory and Charles A. Dana Research Institute, Department of Medicine, Beth Israel Hospital, Harvard Medical School, Boston, MA 02215-5491; tDepartment of Pathology, University of North Carolina, Chapel Hill, NC 27599-7525; and *Laboratory of Experimental Carcinogenesis and Mutagenesis, National Institute of Environmental Health Sciences, Research Triangle Park, NC 27709 Communicated by Seymour J. Klenbanoff, University of Washington School of Medicine, Seattle, WA, July 31, 1996 (received for review April 8, 1996) ABSTRACT Lipid bodies, cytoplasmic inclusions that de- inducible structures. Lipid bodies are prominent in leukocytes velop in cells associated with inflammation, are inducible at sites of natural and experimental inflammation, including structures that might participate in generating inflammatory leukocytes from joints of patients with inflammatory arthritis eicosanoids. Cis-unsaturated fatty acids (arachidonic and (5-7), the airways of patients with acute respiratory distress oleic acids) rapidly induced lipid body formation in leuko- syndrome (8), and casein- or lipopolysaccharide-elicited cytes, and this lipid body induction was inhibited by aspirin guinea pig peritoneal exudates (9). The formation of lipid and nonsteroidal antiinflammatory drugs (NSAIDs). Several bodies is a regulated process that involves activation of intra- findings indicated that the inhibitory effect of aspirin and cellular signaling pathways, including protein kinase C and NSAIDs on lipid body formation was independent of cycloox- phospholipase C (10, 11). -
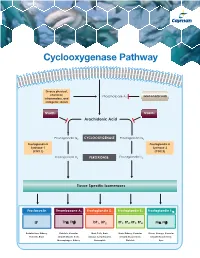
Cyclooxygenase Pathway
Cyclooxygenase Pathway Diverse physical, chemical, Phospholipase A Glucocorticoids inflammatory, and 2 mitogenic stimuli NSAIDs NSAIDs Arachidonic Acid Prostaglandin G2 CYCLOOXYGENASE Prostaglandin G2 Prostaglandin H Prostaglandin H Synthase-1 Synthase-2 (COX 1) (COX 2) Prostaglandin H2 PEROXIDASE Prostaglandin H2 Tissue Specific Isomerases Prostacyclin Thromboxane A2 Prostaglandin D2 Prostaglandin E2 Prostaglandin F2α IP TPα, TPβ DP1, DP2 EP1, EP2, EP3, EP4 FPα, FPβ Endothelium, Kidney, Platelets, Vascular Mast Cells, Brain, Brain, Kidney, Vascular Uterus, Airways, Vascular Platelets, Brain Smooth Muscle Cells, Airways, Lymphocytes, Smooth Muscle Cells, Smooth Muscle Cells, Macrophages, Kidney Eosinophils Platelets Eyes Prostacyclin Item No. Product Features Prostacyclin (Prostaglandin I2; PGI2) is formed from arachidonic acid primarily in the vascular endothelium and renal cortex by sequential 515211 6-keto • Sample Types: Culture Medium | Plasma Prostaglandin • Measure 6-keto PGF levels down to 6 pg/ml activities of COX and prostacyclin synthase. PGI2 is non-enzymatically 1α F ELISA Kit • Incubation : 18 hours | Development: 90-120 minutes | hydrated to 6-keto PGF1α (t½ = 2-3 minutes), and then quickly converted 1α Read: Colorimetric at 405-420 nm to the major metabolite, 2,3-dinor-6-keto PGF1α (t½= 30 minutes). Prostacyclin was once thought to be a circulating hormone that regulated • Assay 24 samples in triplicate or 36 samples in duplicate platelet-vasculature interactions, but the rate of secretion into circulation • NOTE: A portion of urinary 6-keto PGF1α is of renal origin coupled with the short half-life indicate that prostacyclin functions • NOTE : It has been found that normal plasma levels of 6-keto PGF may be low locally. -

Effect of Prostanoids on Human Platelet Function: an Overview
International Journal of Molecular Sciences Review Effect of Prostanoids on Human Platelet Function: An Overview Steffen Braune, Jan-Heiner Küpper and Friedrich Jung * Institute of Biotechnology, Molecular Cell Biology, Brandenburg University of Technology, 01968 Senftenberg, Germany; steff[email protected] (S.B.); [email protected] (J.-H.K.) * Correspondence: [email protected] Received: 23 October 2020; Accepted: 23 November 2020; Published: 27 November 2020 Abstract: Prostanoids are bioactive lipid mediators and take part in many physiological and pathophysiological processes in practically every organ, tissue and cell, including the vascular, renal, gastrointestinal and reproductive systems. In this review, we focus on their influence on platelets, which are key elements in thrombosis and hemostasis. The function of platelets is influenced by mediators in the blood and the vascular wall. Activated platelets aggregate and release bioactive substances, thereby activating further neighbored platelets, which finally can lead to the formation of thrombi. Prostanoids regulate the function of blood platelets by both activating or inhibiting and so are involved in hemostasis. Each prostanoid has a unique activity profile and, thus, a specific profile of action. This article reviews the effects of the following prostanoids: prostaglandin-D2 (PGD2), prostaglandin-E1, -E2 and E3 (PGE1, PGE2, PGE3), prostaglandin F2α (PGF2α), prostacyclin (PGI2) and thromboxane-A2 (TXA2) on platelet activation and aggregation via their respective receptors. Keywords: prostacyclin; thromboxane; prostaglandin; platelets 1. Introduction Hemostasis is a complex process that requires the interplay of multiple physiological pathways. Cellular and molecular mechanisms interact to stop bleedings of injured blood vessels or to seal denuded sub-endothelium with localized clot formation (Figure1). -

Effect of Latanoprost Or 8-Iso Prostaglandin E2 Alone and in Combination on Intraocular Pressure in Glaucomatous Monkey Eyes
LABORATORY SCIENCES Effect of Latanoprost or 8-iso Prostaglandin E2 Alone and in Combination on Intraocular Pressure in Glaucomatous Monkey Eyes Rong-Fang Wang, MD; Steven M. Podos, MD; Janet B. Serle, MD; Thomas W. Mittag, PhD; F. Ventosa, MD; Bernard Becker, MD Objective: To evaluate the possible additivity of the ef- duction of IOP of 4.0 ± 0.6 mm Hg was produced when fects of latanoprost and 8-iso prostaglandin E2 (8-iso PGE2) 8-iso PGE2 was added to latanoprost and of 3.0 ± 0.7 on intraocular pressure (IOP) in monkey eyes with laser- mm Hg was produced when latanoprost was added to 8-iso induced glaucoma. PGE2 on day 13 before the morning dosing. Combina- tion therapy with both agents caused maximum IOP re- Methods: The IOP was measured hourly for 6 hours be- ductions from baseline of 11.3 ± 3.0 mm Hg (33%) (P,.05) ginning at 9:30 AM on day 1 (baseline day), days 6 and 7 (latanoprost with 8-iso PGE2 added) and of 9.8 ± 1.3 , (single-agent therapy), and days 13 and 14 (combination mm Hg (31%) (P .01) (8-iso PGE2 with latanoprost added) therapy with both agents). Following 1 day of baseline mea- on day 14. surement, 4 monkeys with unilateral glaucoma received monotherapy twice daily with either 1 drop of 0.005% la- Conclusion: Latanoprost and 8-iso PGE2 have an addi- tanoprost, or 0.1% 8-iso PGE2, 25 µL, at 9:30 AM and 3:30 tive effect on IOP in glaucomatous monkey eyes.