Americanum Ticks Brian F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ixodes Ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement
International Journal of Molecular Sciences Article Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement Jan Kotál 1,2 , Stéphanie G. I. Polderdijk 3 , Helena Langhansová 1, Monika Ederová 1, Larissa A. Martins 2 , Zuzana Beránková 1, Adéla Chlastáková 1 , OndˇrejHajdušek 4, Michail Kotsyfakis 1,2 , James A. Huntington 3 and JindˇrichChmelaˇr 1,* 1 Department of Medical Biology, Faculty of Science, University of South Bohemia in Ceskˇ é Budˇejovice, Branišovská 1760c, 37005 Ceskˇ é Budˇejovice,Czech Republic; [email protected] (J.K.); [email protected] (H.L.); [email protected] (M.E.); [email protected] (Z.B.); [email protected] (A.C.); [email protected] (M.K.) 2 Laboratory of Genomics and Proteomics of Disease Vectors, Institute of Parasitology, Biology Center CAS, Branišovská 1160/31, 37005 Ceskˇ é Budˇejovice,Czech Republic; [email protected] 3 Cambridge Institute for Medical Research, Department of Haematology, University of Cambridge, The Keith Peters Building, Hills Road, Cambridge CB2 0XY, UK; [email protected] (S.G.I.P.); [email protected] (J.A.H.) 4 Laboratory of Vector Immunology, Institute of Parasitology, Biology Center CAS, Branišovská 1160/31, 37005 Ceskˇ é Budˇejovice,Czech Republic; [email protected] * Correspondence: [email protected] Abstract: Tick saliva is a rich source of antihemostatic, anti-inflammatory, and immunomodulatory molecules that actively help the tick to finish its blood meal. Moreover, these molecules facilitate the Citation: Kotál, J.; Polderdijk, S.G.I.; transmission of tick-borne pathogens. Here we present the functional and structural characterization Langhansová, H.; Ederová, M.; of Iripin-8, a salivary serpin from the tick Ixodes ricinus, a European vector of tick-borne encephalitis Martins, L.A.; Beránková, Z.; and Lyme disease. -

Severe Babesiosis Caused by Babesia Divergens in a Host with Intact Spleen, Russia, 2018 T ⁎ Irina V
Ticks and Tick-borne Diseases 10 (2019) 101262 Contents lists available at ScienceDirect Ticks and Tick-borne Diseases journal homepage: www.elsevier.com/locate/ttbdis Severe babesiosis caused by Babesia divergens in a host with intact spleen, Russia, 2018 T ⁎ Irina V. Kukinaa, Olga P. Zelyaa, , Tatiana M. Guzeevaa, Ludmila S. Karanb, Irina A. Perkovskayac, Nina I. Tymoshenkod, Marina V. Guzeevad a Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russian Federation b Central Research Institute of Epidemiology, Moscow, Russian Federation c Infectious Clinical Hospital №2 of the Moscow Department of Health, Moscow, Russian Federation d Centre for Hygiene and Epidemiology in Moscow, Moscow, Russian Federation ARTICLE INFO ABSTRACT Keywords: We report a case of severe babesiosis caused by the bovine pathogen Babesia divergens with the development of Protozoan parasites multisystem failure in a splenic host. Immunosuppression other than splenectomy can also predispose people to Babesia divergens B. divergens. There was heavy multiple invasion of up to 14 parasites inside the erythrocyte, which had not been Ixodes ricinus previously observed even in asplenic hosts. The piroplasm 18S rRNA sequence from our patient was identical B. Tick-borne disease divergens EU lineage with identity 99.5–100%. Human babesiosis 1. Introduction Leucocyte left shift with immature neutrophils, signs of dysery- thropoiesis, anisocytosis, and poikilocytosis were seen on the peripheral Babesia divergens, a protozoan blood parasite (Apicomplexa: smear. Numerous intra-erythrocytic parasites were found, which were Babesiidae) is primarily specific to bovines. This parasite is widespread initially falsely identified as Plasmodium falciparum. The patient was throughout Europe within the vector Ixodes ricinus. -

Tick-Borne Diseases Primary Tick-Borne Diseases in the Southeastern U.S
Entomology Insect Information Series Providing Leadership in Environmental Entomology Department of Entomology, Soils, and Plant Sciences • 114 Long Hall • Clemson, SC 29634-0315 • Phone: 864-656-3111 email:[email protected] Tick-borne Diseases Primary tick-borne diseases in the southeastern U.S. Affecting Humans in the Southeastern United Disease (causal organism) Tick vector (Scientific name) States Lyme disease Black-legged or “deer” tick (Borrelia burgdorferi species (Ixodes scapularis) Ticks are external parasites that attach themselves complex) to an animal host to take a blood meal at each of Rocky Mountain spotted fever American dog tick their active life stages. Blood feeding by ticks may (Rickettsia rickettsii) (Dermacentor variabilis) lead to the spread of disease. Several common Southern Tick-Associated Rash Lone star tick species of ticks may vector (transmit) disease. Many Illness or STARI (Borrelia (Amblyomma americanum) tick-borne diseases are successfully treated if lonestari (suspected, not symptoms are recognized early. When the disease is confirmed)) Tick-borne Ehrlichiosis not diagnosed during the early stages of infection, HGA-Human granulocytic Black-legged or “deer” tick treatment can be difficult and chronic symptoms anaplasmosis (Anaplasma (Ixodes scapularis) may develop. The most commonly encountered formerly Ehrlichia ticks in the southeastern U.S. are the American dog phagocytophilum) tick, lone star tick, blacklegged or “deer” tick and HME-Human monocytic Lone star tick brown dog tick. While the brown dog tick is notable Ehrlichiosis (Amblyomma americanum) because of large numbers that may be found indoors (Ehrlichia chafeensis ) American dog tick when dogs are present, it only rarely feeds on (Dermacentor variabilis) humans. -

Evidence of Crimean-Congo Haemorrhagic Fever Virus Occurrence in Ixodidae Ticks of Armenia
J Arthropod-Borne Dis, March 2019, 13(1): 9–16 H Gevorgyan et al.: Evidence of Crimean-Congo … Original Article Evidence of Crimean-Congo Haemorrhagic Fever Virus Occurrence in Ixodidae Ticks of Armenia *Hasmik Gevorgyan1,2, Gohar G. Grigoryan1, Hripsime A. Atoyan1, Martin Rukhkyan1, Astghik Hakobyan2, Hovakim Zakaryan3, Sargis A. Aghayan4 1Scientific Center of Zoology and Hydroecology, Yerevan, Armenia 2National Institute of Health, MOH RA, Yerevan, Armenia 3Institute of Molecular Biology NAS RA, Yerevan, Armenia 4Laboratory of Zoology, Research Institute of Biology, Yerevan State University, Yerevan, Armenia (Received 3 Mar 2018; accepted 8 Oct 2018) Abstract Background: Crimean-Congo hemorrhagic fever (CCHF) causes serious health problems in humans. Though ticks of the genera Hyalomma play a significant role in the CCHF virus transmission it was also found in 31 other tick species. Methods: Totally, 1412 ticks from 8 remote sites in Armenia during 2016 were sampled, pooled (3-5 ticks per pool) and tested for the presence of CCHFV antigen using ELISA test. Results: From 359 tick pools, 132 were CCHF virus antigen-positive. From 6 tick species, four species (Rhipicephalus sanguineus, R. annulatus, R. bursa, Hyalomma marginatum) were positive for the virus antigen and R. sanguineus was the most prevalent (37.9%). Dermacentor marginatus and Ixodes ricinus revealed no positive pools, but both revealed delectable but very low virus antigen titers. The highest infection rate (50%) was observed in R. sanguineus, whereas H. marginatus rate of infection was 1 out of 17 pools. Conclusion: For the first time in the last four decades CCHF virus antigen was detected in Ixodid ticks of Armenia. -

Detection of Tick-Borne Pathogens of the Genera Rickettsia, Anaplasma and Francisella in Ixodes Ricinus Ticks in Pomerania (Poland)
pathogens Article Detection of Tick-Borne Pathogens of the Genera Rickettsia, Anaplasma and Francisella in Ixodes ricinus Ticks in Pomerania (Poland) Lucyna Kirczuk 1 , Mariusz Piotrowski 2 and Anna Rymaszewska 2,* 1 Department of Hydrobiology, Faculty of Biology, Institute of Biology, University of Szczecin, Felczaka 3c Street, 71-412 Szczecin, Poland; [email protected] 2 Department of Genetics and Genomics, Faculty of Biology, Institute of Biology, University of Szczecin, Felczaka 3c Street, 71-412 Szczecin, Poland; [email protected] * Correspondence: [email protected] Abstract: Tick-borne pathogens are an important medical and veterinary issue worldwide. Environ- mental monitoring in relation to not only climate change but also globalization is currently essential. The present study aimed to detect tick-borne pathogens of the genera Anaplasma, Rickettsia and Francisella in Ixodes ricinus ticks collected from the natural environment, i.e., recreational areas and pastures used for livestock grazing. A total of 1619 specimens of I. ricinus were collected, including ticks of all life stages (adults, nymphs and larvae). The study was performed using the PCR technique. Diagnostic gene fragments msp2 for Anaplasma, gltA for Rickettsia and tul4 for Francisella were ampli- fied. No Francisella spp. DNA was detected in I. ricinus. DNA of A. phagocytophilum was detected in 0.54% of ticks and Rickettsia spp. in 3.69%. Nucleotide sequence analysis revealed that only one species of Rickettsia, R. helvetica, was present in the studied tick population. The present results are a Citation: Kirczuk, L.; Piotrowski, M.; part of a large-scale analysis aimed at monitoring the level of tick infestation in Northwest Poland. -

Ehrlichiosis and Anaplasmosis
Ehrlichiosis and Anaplasmosis The Diseases and Transmission Ehrlichia and Anaplasma are related bacteria that are transmitted by ticks. These bacteria infect white blood cells in humans. There are three different bacteria that cause disease in humans: Anaplasma phagocytophilum Ehrlichia chaffeensis Ehrlichia ewingii Pathogen (formerly Ehrlichia phagocytophila) Human monocytic Human granulocytic Disease Ehrlichiosis ewingii ehrlichiosis (HME) anaplasmosis (HGA, formerly HGE) Tick Vector Amblyomma americanum (lone star tick) Ixodes scapularis (black-legged tick) Animal reservoirs for E. chaffeensis and E. ewingii are white-tailed deer and dogs. The reservoirs for A. phagocytophilum include cattle, deer, and rodents. You cannot get the diseases directly from animals. The diseases are not spread between humans other than through blood transfusions. Maryland is home to both the lone star tick and the black-legged tick. Symptoms and Treatment Disease Clinical Features . Symptoms appear 7 to 10 days after a tick bite. Symptoms include fever, headache, muscle aches, nausea, vomiting, HME, and loss of appetite. Ehrlichiosis ewingii . Meningoencephalitis occurs in approximately 20% of cases. Development of a rash is possible. This may be confused with Rocky Mountain spotted fever. Symptoms appear 7 to 14 days after a tick bite. HGA . Symptoms include fever, headache, and muscle aches. Meningoencephalitis is rare. Most infections occur when tick activity is highest, in late spring and summer. If left untreated, HME and HGA may be severe. Co-infection with more than one tick borne disease is possible. The immune system is directly infected. Secondary infection and other complications can arise quickly. The elderly and sick are more likely to develop severe illness. -

Australiensis Inducing Mammalian Meat Allergy After Tick Bite
Asia Pac Allergy. 2018 Jul;8(3):e31 https://doi.org/10.5415/apallergy.2018.8.e31 pISSN 2233-8276·eISSN 2233-8268 Educational A novel Australian tick Ixodes & Teaching Material Case Report (Endopalpiger) australiensis inducing mammalian meat allergy after tick bite Mackenzie Kwak1, Colin Somerville2, and Sheryl van Nunen 3,4,* 1Department of Biological Science, National University of Singapore, Singapore 2Allergy West, Perth, WA 6149, Australia 3Tick-induced Allergies Research and Awareness Centre, Chatswood, NSW 2067, Australia 4Northern Clinical School, Sydney Medical School, Faculty of Medicine and Health, The University of Sydney, Sydney, NSW 2006, Australia Received: May 11, 2018 Accepted: Jul 25, 2018 ABSTRACT Tick-induced mammalian meat allergy has become an emergent allergy world-wide after *Correspondence to Sheryl van Nunen van Nunen et al. first described the association between tick bites and the development of Tick-induced Allergies Research and mammalian meat allergy in 2007. Cases of mammalian meat allergy have now been reported Awareness Centre, 40 Johnson Street, on all 6 continents where humans are bitten by ticks, in 17 countries - Australia, United Chatswood, NSW 2067, Australia. States of America (USA), Europe (France, Spain, Germany, Belgium, Switzerland, Sweden, Tel: +61411447365 United Kingdom, Italy, and Norway), Asia (Korea and Japan), Central America (Panama), Fax: +6124121694 South America (Brazil), and Africa (South Africa and Ivory Coast). To date, in each of these E-mail: [email protected] countries, bites from only a single tick species have been linked to the development of Copyright © 2018. Asia Pacific Association of mammalian meat allergy: Ixodes holocyclus (Australia), Amblyomma americanum (USA), Ixodes Allergy, Asthma and Clinical Immunology. -

Tularemia (CFSPH)
Tularemia Importance Tularemia is a zoonotic bacterial disease with a wide host range. Infections are most prevalent among wild mammals and marsupials, with periodic epizootics in Rabbit Fever, lagomorphs and rodents, but clinical cases also occur in sheep, cats and other Deerfly Fever, domesticated species. A variety of syndromes can be seen, but fatal septicemia is Meat-Cutter’s Disease common in some species. In humans, tularemia varies from a localized infection to Ohara Disease, fulminant, life-threatening pneumonia or septicemia. Francis Disease Tularemia is mainly seen in the Northern Hemisphere, where it has recently emerged or re-emerged in some areas, including parts of Europe and the Middle East. A few endemic clinical cases have also been recognized in regions where this disease Last Updated: June 2017 was not thought to exist, such as Australia, South Korea and southern Sudan. In some cases, emergence may be due to increased awareness, surveillance and/or reporting requirements; in others, it has been associated with population explosions of animal reservoir hosts, or with social upheavals such as wars, where sanitation is difficult and infected rodents may contaminate food and water supplies. Occasionally, this disease may even be imported into a country in animals. In 2002, tularemia entered the Czech Republic in a shipment of sick pet prairie dogs from the U.S. Etiology Tularemia is caused by Francisella tularensis (formerly known as Pasteurella tularensis), a Gram negative coccobacillus in the family Francisellaceae and class γ- Proteobacteria. Depending on the author, either three or four subspecies are currently recognized. F. tularensis subsp. tularensis (also known as type A) and F. -
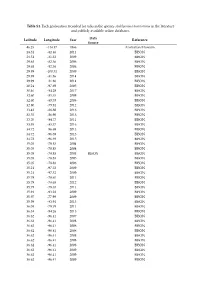
Table S1. Each Geolocation Recorded for Ticks in the Species Amblyomma Americanum in the Literature and Publicly Available Online Databases
Table S1. Each geolocation recorded for ticks in the species Amblyomma americanum in the literature and publicly available online databases. Data Latitude Longitude Year Reference Source 46.25 -114.17 1966 Australian Museum 28.31 -82.46 2011 BISON 28.51 -81.32 2009 BISON 29.68 -82.36 2006 BISON 29.68 -82.36 2006 BISON 29.99 -100.31 2009 BISON 29.99 -81.86 2014 BISON 29.99 -81.86 2014 BISON 30.24 -97.69 2005 BISON 30.46 -84.28 2017 BISON 32.60 -85.35 2008 BISON 32.60 -85.35 2008 BISON 32.80 -79.94 2012 BISON 33.42 -88.88 2016 BISON 33.55 -86.90 2013 BISON 33.70 -84.77 2011 BISON 33.95 -83.37 2016 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2015 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON BISON 35.05 -78.83 2005 BISON 35.05 -78.83 2006 BISON 35.21 -97.32 2009 BISON 35.21 -97.32 2009 BISON 35.79 -78.65 2011 BISON 35.79 -78.65 2012 BISON 35.79 -78.65 2011 BISON 35.91 -93.22 2009 BISON 35.97 -77.99 2009 BISON 35.99 -83.94 2015 BISON 36.08 -79.79 2011 BISON 36.34 -94.26 2015 BISON 36.62 -96.41 2007 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2011 BISON 36.62 -96.41 2012 BISON 36.62 -96.41 2012 BISON 37.36 -77.05 2014 BISON 37.62 -84.87 2016 BISON 37.83 -78.28 2011 BISON 37.97 -85.70 2016 BISON 37.97 -85.70 2016 BISON 38.22 -75.31 2014 BISON 38.43 -88.43 -

Mechanisms Affecting the Acquisition, Persistence and Transmission Of
microorganisms Review Mechanisms Affecting the Acquisition, Persistence and Transmission of Francisella tularensis in Ticks Brenden G. Tully and Jason F. Huntley * Department of Medical Microbiology and Immunology, University of Toledo College of Medicine and Life Sciences, Toledo, OH 43614, USA; [email protected] * Correspondence: [email protected] Received: 29 September 2020; Accepted: 21 October 2020; Published: 23 October 2020 Abstract: Over 600,000 vector-borne disease cases were reported in the United States (U.S.) in the past 13 years, of which more than three-quarters were tick-borne diseases. Although Lyme disease accounts for the majority of tick-borne disease cases in the U.S., tularemia cases have been increasing over the past decade, with >220 cases reported yearly. However, when comparing Borrelia burgdorferi (causative agent of Lyme disease) and Francisella tularensis (causative agent of tularemia), the low infectious dose (<10 bacteria), high morbidity and mortality rates, and potential transmission of tularemia by multiple tick vectors have raised national concerns about future tularemia outbreaks. Despite these concerns, little is known about how F. tularensis is acquired by, persists in, or is transmitted by ticks. Moreover, the role of one or more tick vectors in transmitting F. tularensis to humans remains a major question. Finally, virtually no studies have examined how F. tularensis adapts to life in the tick (vs. the mammalian host), how tick endosymbionts affect F. tularensis infections, or whether other factors (e.g., tick immunity) impact the ability of F. tularensis to infect ticks. This review will assess our current understanding of each of these issues and will offer a framework for future studies, which could help us better understand tularemia and other tick-borne diseases. -

Tularemia – Epidemiology
This first edition of theWHO guidelines on tularaemia is the WHO GUIDELINES ON TULARAEMIA result of an international collaboration, initiated at a WHO meeting WHO GUIDELINES ON in Bath, UK in 2003. The target audience includes clinicians, laboratory personnel, public health workers, veterinarians, and any other person with an interest in zoonoses. Tularaemia Tularaemia is a bacterial zoonotic disease of the northern hemisphere. The bacterium (Francisella tularensis) is highly virulent for humans and a range of animals such as rodents, hares and rabbits. Humans can infect themselves by direct contact with infected animals, by arthropod bites, by ingestion of contaminated water or food, or by inhalation of infective aerosols. There is no human-to-human transmission. In addition to its natural occurrence, F. tularensis evokes great concern as a potential bioterrorism agent. F. tularensis subspecies tularensis is one of the most infectious pathogens known in human medicine. In order to avoid laboratory-associated infection, safety measures are needed and consequently, clinical laboratories do not generally accept specimens for culture. However, since clinical management of cases depends on early recognition, there is an urgent need for diagnostic services. The book provides background information on the disease, describes the current best practices for its diagnosis and treatment in humans, suggests measures to be taken in case of epidemics and provides guidance on how to handle F. tularensis in the laboratory. ISBN 978 92 4 154737 6 WHO EPIDEMIC AND PANDEMIC ALERT AND RESPONSE WHO Guidelines on Tularaemia EPIDEMIC AND PANDEMIC ALERT AND RESPONSE WHO Library Cataloguing-in-Publication Data WHO Guidelines on Tularaemia. -

Tick-Borne Pathogens and Diseases in Greece
microorganisms Review Tick-Borne Pathogens and Diseases in Greece Artemis Efstratiou 1,†, Gabriele Karanis 2 and Panagiotis Karanis 3,4,* 1 National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine, Obihiro 080-8555, Japan; [email protected] 2 Orthopädische Rehabilitationsklinik, Eisenmoorbad Bad Schmiedeberg Kur GmbH, 06905 Bad Schmiedeberg, Germany; [email protected] 3 Medical Faculty and University Hospital, The University of Cologne, 50923 Cologne, Germany 4 Department of Basic and Clinical Sciences, University of Nicosia Medical School, 21 Ilia Papakyriakou, 2414 Engomi. P.O. Box 24005, Nicosia CY-1700, Cyprus * Correspondence: [email protected] † Current address: Max-Planck Institute for Evolutionary Biology, 24306 Plön, Germany. Abstract: Tick-borne diseases (TBDs) are recognized as a serious and growing public health epidemic in Europe, and are a cause of major losses in livestock production worldwide. This review is an attempt to present a summary of results from studies conducted over the last century until the end of the year 2020 regarding ticks, tick-borne pathogens, and tick-borne diseases in Greece. We provide an overview of the tick species found in Greece, as well as the most important tick-borne pathogens (viruses, bacteria, protozoa) and corresponding diseases in circulation. We also consider prevalence data, as well as geographic and climatic conditions. Knowledge of past and current situations of TBDs, as well as an awareness of (risk) factors affecting future developments will help to find approaches to integrated tick management as part of the ‘One Health Concept’; it will assist in avoiding the possibility of hotspot disease emergencies and intra- and intercontinental transmission.