Comparative Study of Hiv, Hcv and Hbv Screening by Third and Fourth Generation Elisa in Donated Blood
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Chennai Comprehensive Transportation Study (CCTS)
ACKNOWLEDGEMENT The consultants are grateful to Tmt. Susan Mathew, I.A.S., Addl. Chief Secretary to Govt. & Vice-Chairperson, CMDA and Thiru Dayanand Kataria, I.A.S., Member - Secretary, CMDA for the valuable support and encouragement extended to the Study. Our thanks are also due to the former Vice-Chairman, Thiru T.R. Srinivasan, I.A.S., (Retd.) and former Member-Secretary Thiru Md. Nasimuddin, I.A.S. for having given an opportunity to undertake the Chennai Comprehensive Transportation Study. The consultants also thank Thiru.Vikram Kapur, I.A.S. for the guidance and encouragement given in taking the Study forward. We place our record of sincere gratitude to the Project Management Unit of TNUDP-III in CMDA, comprising Thiru K. Kumar, Chief Planner, Thiru M. Sivashanmugam, Senior Planner, & Tmt. R. Meena, Assistant Planner for their unstinted and valuable contribution throughout the assignment. We thank Thiru C. Palanivelu, Member-Chief Planner for the guidance and support extended. The comments and suggestions of the World Bank on the stage reports are duly acknowledged. The consultants are thankful to the Steering Committee comprising the Secretaries to Govt., and Heads of Departments concerned with urban transport, chaired by Vice- Chairperson, CMDA and the Technical Committee chaired by the Chief Planner, CMDA and represented by Department of Highways, Southern Railways, Metropolitan Transport Corporation, Chennai Municipal Corporation, Chennai Port Trust, Chennai Traffic Police, Chennai Sub-urban Police, Commissionerate of Municipal Administration, IIT-Madras and the representatives of NGOs. The consultants place on record the support and cooperation extended by the officers and staff of CMDA and various project implementing organizations and the residents of Chennai, without whom the study would not have been successful. -
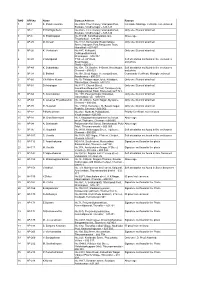
SNO APP.No Name Contact Address Reason 1 AP-1 K
SNO APP.No Name Contact Address Reason 1 AP-1 K. Pandeeswaran No.2/545, Then Colony, Vilampatti Post, Intercaste Marriage certificate not enclosed Sivakasi, Virudhunagar – 626 124 2 AP-2 P. Karthigai Selvi No.2/545, Then Colony, Vilampatti Post, Only one ID proof attached. Sivakasi, Virudhunagar – 626 124 3 AP-8 N. Esakkiappan No.37/45E, Nandhagopalapuram, Above age Thoothukudi – 628 002. 4 AP-25 M. Dinesh No.4/133, Kothamalai Road,Vadaku Only one ID proof attached. Street,Vadugam Post,Rasipuram Taluk, Namakkal – 637 407. 5 AP-26 K. Venkatesh No.4/47, Kettupatti, Only one ID proof attached. Dokkupodhanahalli, Dharmapuri – 636 807. 6 AP-28 P. Manipandi 1stStreet, 24thWard, Self attestation not found in the enclosures Sivaji Nagar, and photo Theni – 625 531. 7 AP-49 K. Sobanbabu No.10/4, T.K.Garden, 3rdStreet, Korukkupet, Self attestation not found in the enclosures Chennai – 600 021. and photo 8 AP-58 S. Barkavi No.168, Sivaji Nagar, Veerampattinam, Community Certificate Wrongly enclosed Pondicherry – 605 007. 9 AP-60 V.A.Kishor Kumar No.19, Thilagar nagar, Ist st, Kaladipet, Only one ID proof attached. Thiruvottiyur, Chennai -600 019 10 AP-61 D.Anbalagan No.8/171, Church Street, Only one ID proof attached. Komathimuthupuram Post, Panaiyoor(via) Changarankovil Taluk, Tirunelveli, 627 761. 11 AP-64 S. Arun kannan No. 15D, Poonga Nagar, Kaladipet, Only one ID proof attached. Thiruvottiyur, Ch – 600 019 12 AP-69 K. Lavanya Priyadharshini No, 35, A Block, Nochi Nagar, Mylapore, Only one ID proof attached. Chennai – 600 004 13 AP-70 G. -

Tamil Nadu Government Gazette
© [Regd. No. TN/CCN/467/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 197/2009. 2013 [Price: Rs. 54.80 Paise. TAMIL NADU GOVERNMENT GAZETTE PUBLISHED BY AUTHORITY No. 41] CHENNAI, WEDNESDAY, OCTOBER 23, 2013 Aippasi 6, Vijaya, Thiruvalluvar Aandu–2044 Part VI—Section 4 Advertisements by private individuals and private institutions CONTENTS PRIVATE ADVERTISEMENTS Pages Change of Names .. 2893-3026 Notice .. 3026-3028 NOTICE NO LEGAL RESPONSIBILITY IS ACCEPTED FOR THE PUBLICATION OF ADVERTISEMENTS REGARDING CHANGE OF NAME IN THE TAMIL NADU GOVERNMENT GAZETTE. PERSONS NOTIFYING THE CHANGES WILL REMAIN SOLELY RESPONSIBLE FOR THE LEGAL CONSEQUENCES AND ALSO FOR ANY OTHER MISREPRESENTATION, ETC. (By Order) Director of Stationery and Printing. CHANGE OF NAMES 43888. My son, D. Ramkumar, born on 21st October 1997 43891. My son, S. Antony Thommai Anslam, born on (native district: Madurai), residing at No. 4/81C, Lakshmi 20th March 1999 (native district: Thoothukkudi), residing at Mill, West Colony, Kovilpatti, Thoothukkudi-628 502, shall Old No. 91/2, New No. 122, S.S. Manickapuram, Thoothukkudi henceforth be known as D. RAAMKUMAR. Town and Taluk, Thoothukkudi-628 001, shall henceforth be G. DHAMODARACHAMY. known as S. ANSLAM. Thoothukkudi, 7th October 2013. (Father.) M. v¯ð¡. Thoothukkudi, 7th October 2013. (Father.) 43889. I, S. Salma Banu, wife of Thiru S. Shahul Hameed, born on 13th September 1975 (native district: Mumbai), 43892. My son, G. Sanjay Somasundaram, born residing at No. 184/16, North Car Street, on 4th July 1997 (native district: Theni), residing Vickiramasingapuram, Tirunelveli-627 425, shall henceforth at No. 1/190-1, Vasu Nagar 1st Street, Bank be known as S SALMA. -

University of Madras Institute of Distance Education Centre Notification May – June 2012 Examinations Chennai City Centres (Ug Degree Courses)
UNIVERSITY OF MADRAS INSTITUTE OF DISTANCE EDUCATION CENTRE NOTIFICATION MAY – JUNE 2012 EXAMINATIONS CHENNAI CITY CENTRES (UG DEGREE COURSES) S. No. CENTRE / VENUE EXAMINATIONS 1. J.H.A. Agarsan College, Manjambakkam B.Com – A11 and C11 Batches (Men and Road, Madhavaram, Chennai – 600 060. Women) Phone: 25559001 / 25559004 2. Thiruthangal Nadar College, Selavayal, B.Com – A10 & C10 Batches (All Chennai – 600051 (Near Kaviarasu Candidates Men & Women) Kannadasan Nagar Bus Depot) Phone: 25941717, 25942525, 25940393. 3. King’s Matriculation Higher Secondary B.Com – A9 & C9 Batches (Men and School, 85, Villivakkam Road, Women) Lakshmipuram Kalpalayam, Chennai – 600099. Phone: 25655709 / 25650348 4. I.D.P.L. Matric Higher Secondary School, B.Com – A7, A8, C7 & C8, X5 & X6 IDPL Township, Nandambakkam, Chennai Batches (All candidates Men & Women) – 600089. B.Sc. Geography – All Candidates Men Phone:22345534 & Women) 5. Shri Krishnaswamy College for Women, B.Com Old Batches up to X4 UCM / AC-48, 6th Main Road, Anna Nagar, SCM – All candidates Men & Women Chennai – 600040. Phone: 26282977 Bachelor of Multimedia and Animation (BMM) (All Batches Men and Women) 6. Alpha Arts and Science College, 16, B.B.A. – A11 & C11 Batches – (All College Road, Thundalam, Chennai – Candidates Men & Women) 600116. Phone: 24768656 7. Annai Violet Arts and Science College, B.B.A. – A10 & C10 Batches – (All No.53, Menambedu, Ambattur, Chennai – Candidates Men & Women) 600053. Phone: 26861611 / 26864684 B.A. Yoga for Human Excellence – (All Batches Men & Women) 8. Bhaktavatsalam Memorial College for BBA – A9 & C9 Batches – (Men and Women, No.14, 31st Street, Periyar Nagar, Women) Korattur, Chennai – 600 080. Phone: 26872891 9. -

Summary of Family Membership and Gender by Club MBR0018 As of February, 2010
Summary of Family Membership and Gender by Club MBR0018 as of February, 2010 Club Fam. Unit Fam. Unit Club Ttl. Club Ttl. District Number Club Name HH's 1/2 Dues Females Male TOTAL District 324A6 26412 AVADI 5 5 6 49 55 District 324A6 26418 ENNORE TIRUVOTTIYUR 0 0 2 8 10 District 324A6 26438 CHENNAI SHANTHI COLONY 3 3 3 11 14 District 324A6 26449 TAMBARAM 1 1 0 19 19 District 324A6 29705 VILLAVAKKAM 3 3 4 18 22 District 324A6 32179 PALLAVAPURAM 6 6 6 39 45 District 324A6 32675 TAMBARAM WEST 1 0 5 27 32 District 324A6 35460 MADRAS METROPOLITAN SO 5 5 6 12 18 District 324A6 36531 MADRAS RED HILLS 25 26 27 90 117 District 324A6 37537 MADRAS METRO EAST 48 48 55 80 135 District 324A6 40288 MADRAS SEMBIUM 2 2 2 21 23 District 324A6 45544 MADRAS SELAIYUR 0 0 0 18 18 District 324A6 49225 MADRAS TRIDENT PALLAVARAM 0 0 0 19 19 District 324A6 51650 MADRAS PERIPHERAL CITY 8 9 7 33 40 District 324A6 54882 MADRAS ARUMBAKKAM 0 0 0 12 12 District 324A6 55709 MADRAS KOLATHOOR 0 0 0 8 8 District 324A6 57011 CHITLAPAKKAM 0 0 1 39 40 District 324A6 60194 MADRAS HASTHINAPURAM 4 4 6 11 17 District 324A6 60920 CHENNAI AMBASSADORS 11 11 11 22 33 District 324A6 61600 MADRAS MADIPAKKAM 0 0 0 16 16 District 324A6 62049 MADRAS BHARATHAM-PERAVALLUR 0 0 1 15 16 District 324A6 62244 CHENNAI TIRUVOTTIYUR EAST 1 1 1 23 24 District 324A6 62543 MADRAS TAMBARAM EAST 4 5 9 16 25 District 324A6 63671 CHENNAI MADAMBAKKAM 0 0 0 23 23 District 324A6 63681 CHENNAI AGARAM 0 0 0 26 26 District 324A6 63701 CHENNAI GUMMIDIPOONDI 1 1 1 30 31 District 324A6 64330 CHENNAI KUNRATHUR -

Tamil Nadu Government Gazette
© [Regd. No. TN/CCN/467/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 197/2009. 2012 [Price: Rs. 28.80 Paise. TAMIL NADU GOVERNMENT GAZETTE PUBLISHED BY AUTHORITY No. 37] CHENNAI, WEDNESDAY, SEPTEMBER 19, 2012 Purattasi 3, Thiruvalluvar Aandu–2043 Part VI—Section 4 Advertisements by private individuals and private institutions CONTENTS PRIVATE ADVERTISEMENTS Change of Names .. 2325-2395 Notices .. 2240-2242 .. 1764 1541-1617 NoticeNOTICE .. 1617 NO LEGAL RESPONSIBILITY IS ACCEPTED FOR THE PUBLICATION OF ADVERTISEMENTS REGARDING CHANGE OF NAME IN THE TAMIL NADU GOVERNMENT GAZETTE. PERSONS NOTIFYING THE CHANGES WILL REMAIN SOLELY RESPONSIBLE FOR THE LEGAL CONSEQUENCES AND ALSO FOR ANY OTHER MISREPRESENTATION, ETC. (By Order) Director of Stationery and Printing. CHANGE OF NAMES 34603. My daughter, N. Rizwana, born on 12th November 34606. I, B. Vel, son of Thiru K. Bethuraj, born on 4th April 1996 (native district: Ramanathapuram), residing at 1967 (native district: Theni), residing at No. 63-NA, Vaigai No. 11-7B/68, New Ramnad Road, Visalam, Madurai- Colony, Anna Nagar, Madurai-625 020, shall henceforth be 625 009, shall henceforth be known as N. RISVANA BEGAM. known as B. VELUCHAMY. ï£Ã˜H„¬ê. B. VEL. Madurai, 10th September 2012. (Father.) Madurai, 10th September 2012. 34604. I, N. Radha, daughter of Thiru V. Navaneethan, 34607. I, N. Thirupathi, son of Thiru Narayanasamy, born born on 18th July 1991 (native district: Calcutta-West Bengal), on 7th June 1983 (native district: Dindigul), residing at residing at No. 1/65A, Katchaikatti, T. Vadipatti Taluk, Madurai-625 218, shall henceforth be known No. 1C, Melaponnakaram 2nd Street, Arapalayam Post, as N. -

Storm Water Flood Modeling in the Sub- Basin of Chennai Corporation, Chennai, Tamilnadu, India
Storm Water Flood Modeling in the Sub- basin of Chennai Corporation, Chennai, Tamilnadu, India Dr. Y. R. Satyaji Rao Scientist F and Head Deltaic Regional Centre National Institute of Hydrology Kakinada 533003, Andhra Pradesh, India Website: www.nih.ernet.in Types of Flooding (Geographically): Riverine flooding It happens when extreme rainfall attacks in a river basin (Mississippi, 1993; Miller, 1997; Changman, 1998; Li and Guo et al., 1999; NVE, 2000; Meade, 2002). Urban flooding It is triggered when surface runoff exceeds the capacity of drainage systems, which happens when heavy rainfall pours on sewers with the limited capacity, or even medium rainfall falls on poorly planned or operated drainage systems (Kamal and Rabbi, 1998; Arambepola, 2002). Coastal flooding It takes place when heavy rainfall on inland encounters storm surges from the sea (Miller, 1997; Barry, 1997; Smith and Ward, 1998; Parker1, 2000; Pilarczyk and Nuoi, 2002). Definition of Urban Drainage Systems Urban drainage systems are defined as physical facilities that collect, store, convey, and treat runoff in urban areas. These facilities normally include detention and retention facilities, streets, storm sewers, inlets, open channels, and special structures such as inlets, manholes, and energy dissipaters” (ASCE and WEF, 1992). Why Urban Floods are increasing ? Increase in Flood peak and storm water network designs are old (Design limitations) Improper maintenance of storm water network (Carrying capacity) Impact of Boundary Conditions (Confluence points/backwater/tides) -

B29N Bus Time Schedule & Line Route
B29N bus time schedule & line map B29N Otteri View In Website Mode The B29N bus line (Otteri) has 4 routes. For regular weekdays, their operation hours are: (1) Otteri: 8:55 PM (2) Perambur B.S.: 10:00 AM (3) Periyar Nagar: 6:58 PM (4) Velachery: 8:10 AM - 5:28 PM Use the Moovit App to ƒnd the closest B29N bus station near you and ƒnd out when is the next B29N bus arriving. Direction: Otteri B29N bus Time Schedule 16 stops Otteri Route Timetable: VIEW LINE SCHEDULE Sunday 8:55 PM Monday 8:55 PM Periyar Nagar Tuesday 8:55 PM S.R.B Colony Wednesday 8:55 PM 70 Feet Road Thursday 8:55 PM Agaram Junction Friday 8:55 PM Kamarajar Salai Saturday 8:55 PM Veenus Chembium Old Post O∆ce B29N bus Info Lourde School Direction: Otteri Stops: 16 Trip Duration: 11 min Mangalapuri Line Summary: Periyar Nagar, S.R.B Colony, 70 Feet Chandra Yogi Samadhi Rd, Chennai Road, Agaram Junction, Kamarajar Salai, Veenus, Chembium Old Post O∆ce, Lourde School, Perambur Mangalapuri, Perambur, Jamalaya, Mettupalayam, Otteri Church, Binny Mills, Otteri Police Station, Otteri Jamalaya Mettupalayam Otteri Church Binny Mills Otteri Police Station Otteri Kancheepuram, Chennai Otteri Otteri Bridge, Chennai Direction: Perambur B.S. B29N bus Time Schedule 56 stops Perambur B.S. Route Timetable: VIEW LINE SCHEDULE Sunday 10:00 AM Monday 10:00 AM Velachery (Vijaynagar) Velachery Bypass Road, Chennai Tuesday 10:00 AM Velachery (Vijaynagar) Wednesday 10:00 AM Bus Way, Chennai Thursday 10:00 AM Dhandeswaram Friday 10:00 AM Thandeeswaram Saturday 10:00 AM Gandhi Nagar Gurunanak College Velachery Main Road, Chennai B29N bus Info Direction: Perambur B.S. -

Macro Drainage System in Cma
Chapter X MACRO DRAINAGE SYSTEM IN CMA Introduction: CMA lies along the east coast of Southern India and is traversed by three major rivers namely Kosasthalaiyar River, Cooum River and Adyar River. The climate of the region is dominated by the monsoons, which are caused by thermal contrast between land and sea. Monsoon climates are characterised by clearly marked seasons with specific types of wind and weather. The South West monsoon dominates weather patterns in Tamilnadu from July –September and is characterised by periods of sultry wet weather. Rain shadow effects limit rainfall in the east coast in Tamilnadu and it is light or intermittent during this season. This period is followed by North-East Monsoon, which brings cool cloudy weather, relatively free of rain over most of the monsoon- dominated land (India). The exception is South-East-India including Tamilnadu where about 78% of the annual rainfall occurs at this time. The start of the heavy rains usually falls in October lasting up to December. Most of the rainfall is associated with clear synoptic systems of depressions and cyclones with night time rainfall most common. In CMA between October and December most of the rainfall occurs and it is rare between January and April. 10.02 River Nagari which has a large catchment area in the Chittoor District (Andhra Pradesh) region and the Nandi River, which has catchment area in the Vellore District, join near Kanakamma Chattiram and enter Poondi Reservoir. Kosasthalaiyar River, which has its origin near Kaveripakkam and has catchment area in North Arcot District, has a branch near Kesavaram Anicut and flows to the city as Cooum River and the main Kosasthalaiyar river flows to Poondi reservoir. -

25321180 / 26415700 E-Mail : [email protected] Date: 01.08.2019 E-AUCTION SALE NOTICE
INDIAN OVERSEAS BANK PURASAWALKAM BRANCH 15 Hunters Road, Purasawalkam, Chennai - 600 112. Ph : 25321180 / 26415700 E-mail : [email protected] Date: 01.08.2019 E-AUCTION SALE NOTICE SALE OF IMMOVABLE PROPERTY MORTGAGED TO THE BANK UNDER THE SECURITISATION AND RECONSTRUCTION OF FINANCIAL ASSETS AND ENFORCEMENT OF SECURITY INTEREST ACT, 2002 Whereas M/s Shapes & Shades Retail Pvt. Ltd. Represented by its Directors 1.Mr. Dulichand Lodha, 2.Mrs. Kanchana Devi Lodha, 3.Mr. Padam Chand Lodha,and 4. Mrs. V M Latha having Registered address at No.49, Audiappa Naicken Street, Sowcarpet Chennai – 600 079 has borrowed monies from Indian Overseas Bank against the mortgage of the immovable properties more fully described in the schedule hereunder and on upon classification of the account as NPA, the Bank has issued a demand notice under Section 13(2) of the SARFAESI Act, 2002 (Act) on 07.06.2018 calling upon the borrowers M/s Shapes & Shades Retail Pvt. Ltd. Represented by its Directors 1.Mr. Dulichand Lodha, 2.Mrs. Kanchana Devi Lodha, 3.Mr. Padam Chand Lodha,and 4. Mrs. V M Latha having Registered address at No.49, Audiappa Naicken Street, Sowcarpet Chennai – 600 079 to pay the amount due to the Bank, being Rs.5,15,12,703/- (Rupees Five Crore Fifteen Lakhs Twelve Thousand Seven Hundred and Three only) as on 06.06.2018 payable together with further interest at contractual rates and rests along with costs, charges etc till date of repayment within 60 days from the date of receipt of the said notice. Whereas the borrowers & guarantors having failed to pay the amount dues in full to the Bank as called for in the said demand notice, the Bank has taken symbolic possession of the secured assets more fully described in the schedule hereunder on 29.10.2018 under Section 13 (4) of the Act with the right to sell the same in “As is where is” and “As is what is” basis under Section13(4) of the Act read with Rules 8 &9 of the Security interest (Enforcement) Rules, 2002 for realization of Bank’s dues. -
Short Term Course on “Teaching and Learning Techniques in Higher Education” (From 21.07.2014 to 26.07.2014)
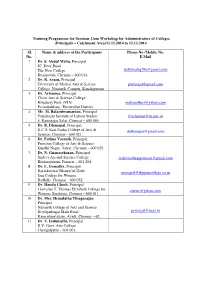
Training Programme for Seminar Cum Workshop for Administrators of Colleges (Principals – Catchment Area)11.12.2014 to 12.12.2014 Sl. Name & address of the Participants Phone No /Mobile No. No. E.Mail 1 Dr. S. Abdul Maliq , Principal 87, Peter Road The New College [email protected] Royapettah, Chennai – 600 014. 2 Dr. R. Arasu , Principal University of Madras Arts & Science [email protected] College, Nemmeli Campus, Kanchipuram 3 Dr. Arlamma , Principal Christ Arts & Science College Kilachery Post, (VIA) [email protected] Perambakkam, Thiruvallur District 4 Mr. M. Balasubramanian , Principal Tamilnadu Institute of Labour Studies [email protected] 5, Kamarajar Salai, Chennai – 600 005. 5 Dr. R. Dhanapal , Principal K.C.S. Kasi Nadar College of Arts & [email protected] Science, Chennai – 600 021. 6 Dr. Fatima Vasanth , Principal Patrician College of Arts & Science Gandhi Nagar, Adyar, Chennai – 600 020. 7 Dr. N. Gnanasekaran , Principal Sridevi Ars and Science College [email protected] Krishnapuram, Ponneri – 601 204. 8 Dr. L. Gomathy , Principal Ratankanwar Bhawarlal Gothi [email protected] Jain College for Women Redhills, Chennai – 600 052. 9 Dr. Hanifa Ghosh , Principal Chevalier T. Thomas Elizabeth College for [email protected] Women, Sembiam, Chennai – 600 011. 10 Dr. Mrs. Hemalatha Thiagarajan Principal Nazareth College of Arts and Science Kovilpathagai Main Road [email protected] Kannadapalayam, Avadi, Chennai – 62. 11 Dr. V. Indumathi , Principal R.V. Govt. Arts College Chengalpattu – 603 001. 12 Dr. C. Joseph Barnabas , Principal Hindustan College of Arts & Science [email protected] Rajiv Gandhi Road (OMR) Kelambakkam, Chennai – 603 103. 13 Rev. Dr. G. Joseph Antony Samy, S.J. -

Mint Building S.O Chennai TAMIL NADU
pincode officename districtname statename 600001 Flower Bazaar S.O Chennai TAMIL NADU 600001 Chennai G.P.O. Chennai TAMIL NADU 600001 Govt Stanley Hospital S.O Chennai TAMIL NADU 600001 Mannady S.O (Chennai) Chennai TAMIL NADU 600001 Mint Building S.O Chennai TAMIL NADU 600001 Sowcarpet S.O Chennai TAMIL NADU 600002 Anna Road H.O Chennai TAMIL NADU 600002 Chintadripet S.O Chennai TAMIL NADU 600002 Madras Electricity System S.O Chennai TAMIL NADU 600003 Park Town H.O Chennai TAMIL NADU 600003 Edapalayam S.O Chennai TAMIL NADU 600003 Madras Medical College S.O Chennai TAMIL NADU 600003 Ripon Buildings S.O Chennai TAMIL NADU 600004 Mandaveli S.O Chennai TAMIL NADU 600004 Vivekananda College Madras S.O Chennai TAMIL NADU 600004 Mylapore H.O Chennai TAMIL NADU 600005 Tiruvallikkeni S.O Chennai TAMIL NADU 600005 Chepauk S.O Chennai TAMIL NADU 600005 Madras University S.O Chennai TAMIL NADU 600005 Parthasarathy Koil S.O Chennai TAMIL NADU 600006 Greams Road S.O Chennai TAMIL NADU 600006 DPI S.O Chennai TAMIL NADU 600006 Shastri Bhavan S.O Chennai TAMIL NADU 600006 Teynampet West S.O Chennai TAMIL NADU 600007 Vepery S.O Chennai TAMIL NADU 600008 Ethiraj Salai S.O Chennai TAMIL NADU 600008 Egmore S.O Chennai TAMIL NADU 600008 Egmore ND S.O Chennai TAMIL NADU 600009 Fort St George S.O Chennai TAMIL NADU 600010 Kilpauk S.O Chennai TAMIL NADU 600010 Kilpauk Medical College S.O Chennai TAMIL NADU 600011 Perambur S.O Chennai TAMIL NADU 600011 Perambur North S.O Chennai TAMIL NADU 600011 Sembiam S.O Chennai TAMIL NADU 600012 Perambur Barracks S.O Chennai