The Contribution of the Ankyrin Repeat Domain of TRPV1 As a Thermal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ankrd9 Is a Metabolically-Controlled Regulator of Impdh2 Abundance and Macro-Assembly
ANKRD9 IS A METABOLICALLY-CONTROLLED REGULATOR OF IMPDH2 ABUNDANCE AND MACRO-ASSEMBLY by Dawn Hayward A dissertation submitted to The Johns Hopkins University in conformity with the requirements of the degree of Doctor of Philosophy Baltimore, Maryland April 2019 ABSTRACT Members of a large family of Ankyrin Repeat Domains proteins (ANKRD) regulate numerous cellular processes by binding and changing properties of specific protein targets. We show that interactions with a target protein and the functional outcomes can be markedly altered by cells’ metabolic state. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 by binding to vesicles. Upon nutrient limitation, ANKRD9 loses association with vesicles and assembles with IMPDH2 into rod-like structures, in which IMPDH2 is stable. Inhibition of IMPDH2 with Ribavirin favors ANKRD9 binding to rods. The IMPDH2/ANKRD9 assembly is reversed by guanosine, which restores association of ANKRD9 with vesicles. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicles-to-rods transition as well as binding and regulation of IMPDH2. ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Thus, the status of guanosine pools affects the mode of ANKRD9 action towards IMPDH2. Advisor: Dr. Svetlana Lutsenko, Department of Physiology, Johns Hopkins University School of Medicine Second reader: -

Repeat Proteins Challenge the Concept of Structural Domains
844 Biochemical Society Transactions (2015) Volume 43, part 5 Repeat proteins challenge the concept of structural domains Rocıo´ Espada*1, R. Gonzalo Parra*1, Manfred J. Sippl†, Thierry Mora‡, Aleksandra M. Walczak§ and Diego U. Ferreiro*2 *Protein Physiology Lab, Dep de Qu´ımica Biologica, ´ Facultad de Ciencias Exactas y Naturales, UBA-CONICET-IQUIBICEN, Buenos Aires, C1430EGA, Argentina †Center of Applied Molecular Engineering, Division of Bioinformatics, Department of Molecular Biology, University of Salzburg, 5020 Salzburg, Austria ‡Laboratoire de physique statistique, CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France §Laboratoire de physique theorique, ´ CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France Abstract Structural domains are believed to be modules within proteins that can fold and function independently. Some proteins show tandem repetitions of apparent modular structure that do not fold independently, but rather co-operate in stabilizing structural forms that comprise several repeat-units. For many natural repeat-proteins, it has been shown that weak energetic links between repeats lead to the breakdown of co-operativity and the appearance of folding sub-domains within an apparently regular repeat array. The quasi-1D architecture of repeat-proteins is crucial in detailing how the local energetic balances can modulate the folding dynamics of these proteins, which can be related to the physiological behaviour of these ubiquitous biological systems. Introduction and between repeats, challenging the concept of structural It was early on noted that many natural proteins typically domain. collapse stretches of amino acid chains into compact units, defining structural domains [1]. These domains typically correlate with biological activities and many modern proteins can be described as composed by novel ‘domain arrange- Definition of the repeat-units ments’ [2]. -

A Structural Guide to Proteins of the NF-Kb Signaling Module
Downloaded from http://cshperspectives.cshlp.org/ on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press A Structural Guide to Proteins of the NF-kB Signaling Module Tom Huxford1 and Gourisankar Ghosh2 1Department of Chemistry and Biochemistry, San Diego State University, 5500 Campanile Drive, San Diego, California 92182-1030 2Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0375 Correspondence: [email protected] The prosurvival transcription factor NF-kB specifically binds promoter DNA to activate target gene expression. NF-kB is regulated through interactions with IkB inhibitor proteins. Active proteolysis of these IkB proteins is, in turn, under the control of the IkB kinase complex (IKK). Together, these three molecules form the NF-kB signaling module. Studies aimed at charac- terizing the molecular mechanisms of NF-kB, IkB, and IKK in terms of their three-dimen- sional structures have lead to a greater understanding of this vital transcription factor system. F-kB is a master transcription factor that from the perspective of their three-dimensional Nresponds to diverse cell stimuli by activat- structures. ing the expression of stress response genes. Multiple signals, including cytokines, growth factors, engagement of the T-cell receptor, and NF-kB bacterial and viral products, induce NF-kB Introduction to NF-kB transcriptional activity (Hayden and Ghosh 2008). A point of convergence for the myriad NF-kB was discovered in the laboratory of of NF-kB inducing signals is the IkB kinase David Baltimore as a nuclear activity with bind- complex (IKK). Active IKK in turn controls ing specificity toward a ten-base-pair DNA transcription factor NF-kB by regulating pro- sequence 50-GGGACTTTCC-30 present within teolysis of the IkB inhibitor protein (Fig. -

Allicin Protects Against Lipopolysaccharide-Induced Acute Lung Injury by Up-Regulation of Claudin-4
Zheng et al Tropical Journal of Pharmaceutical Research July 2014; 13 (7): 1063-1069 ISSN: 1596-5996 (print); 1596-9827 (electronic) © Pharmacotherapy Group, Faculty of Pharmacy, University of Benin, Benin City, 300001 Nigeria. All rights reserved. Available online at http://www.tjpr.org http://dx.doi.org/10.4314/tjpr.v13i7.8 Original Research Article Allicin Protects against Lipopolysaccharide-Induced Acute Lung Injury by Up-Regulation of Claudin-4 Yue-liang Zheng, Wen-wei Cai, Guang-zhao Yan, Yuan-zhan Xu and Mei-qi Zhang* Department of Emergency, Zhejiang Provincial People's Hospital, Hangzhou 310014, China *For correspondence: Email: [email protected]; Tel: +86-0571-85893631 Received: 8 January 2014 Revised accepted: 31 May 2014 Abstract Purpose: To investigate the effect of allicin, an active component of garlic, on lipopolysaccharide (LPS)- induced acute lung injury. Methods: Wistar rats were subjected to LPS intravenous injection with or without allicin treatment to induce acute lung injury (ALI) model. Also, A549 cells were stimulated with LPS in the presence and absence of allicin. HE staining was used to detect pathological changes in lung tissues. Enzyme-linked immunosorbent assay (ELISA) was performed to measure cytokine content. Cell viability was measured by CCK-8 and EdU incorporation assay. Genes expression was determined by real time polymerase chain reaction (PCR) and Western blot. Flow cytometry was applied to measure cell apoptosis. Results: In vivo data showed that pulmonary edema, inflammatory cytokines expression and pathological changes were significantly attenuated in LPS-induced ALI after treatment with allicin (p < 0.05) while in vitro results indicate that allicin administration significantly improved the A549 cell viability in a dose-dependent manner as measured by CCK-8 and EdU incorporation assay. -
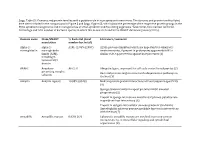
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

World Journal of Pharmaceutical Research Anjna Et Al
World Journal of Pharmaceutical Research Anjna et al. World Journal of PharmaceuticalSJIF ImpactResearch Factor 8.074 Volume 8, Issue 10, 741-748. Research Article ISSN 2277– 7105 A PHYTOCHEMICAL STUDY OF ALLIUM SATIVUM – W.S.R TO ALLICIN CONTENT 1*Tak Anjna, 2Thakur Kumar Sudarshan and 3Das Kumar Arun 1Associate Professor, Prasuti Tantra Avum Stri Roga, Main Campus, Uttrakhanda Ayurveda University, Dehradun, Uttarakhanda. 2Lecturer, Ras Shashtra Avum Bhaishajya Kalpana, R.G.G.P.G. Ayurvedic College, Paprola, Himachal Pradesh. 3Principal, Professor & H.O.D, Ras Shashtra Avum Bhaishajya Kalpana, Govt. Ayurvedic College, Bolangir, Orisa. ABSTRACT Article Received on 15 July 2019, Allium sativum has attracted the interest of many researchers due to its Revised on 05 August 2019, wide range of therapeutic effects with minimal adverse reactions. Its Accepted on 25 August 2019, DOI: 10.20959/wjpr201910-15253 role in promoting the female reproductive health can be well understood from the fact Acharya Kashyap in his text has described a full chapter Lashuna Kalpadhyaya mentioning that the woman *Corresponding Author Tak Anjna consuming Lashuna will not suffer from diseases of kati, shroni Associate Professor, Prasuti (pelvis), gramyadharma janya rogas (sexually transmitted diseases) Tantra Avum Stri Roga, and infertility. Its effects are mainly attributed to its chemical Main Campus, Uttrakhanda constituents like Allicin, Ajoene and certain other sulphur compounds Ayurveda University, etc. In the present study, bulbs of Allium sativum were dried and in Dehradun, Uttarakhanda. controlled temperature and fine powder was made. It was filled in capsules and clinical trial was done in the patients of Hypomenorrhoea. In context of this, a phytochemical study of dried powder of Allium sativum was done and various chemical constituents of garlic have been investigated to support its pharmaco-therapeutic actions as per clinical study. -

Allicin Induces Apoptosis Through Activation of Both Intrinsic and Extrinsic Pathways in Glioma Cells
5976 MOLECULAR MEDICINE REPORTS 17: 5976-5981, 2018 Allicin induces apoptosis through activation of both intrinsic and extrinsic pathways in glioma cells CHENLONG LI1, HANGUANG JING2, GUANGTAO MA3 and PENG LIANG1 1Department of Neurosurgery, Harbin Medical University Cancer Hospital, Harbin, Heilongjiang 150086; 2Basic Theory of Chinese Medicine, Preclinical Medicine School, Beijing University of Chinese Medicine, Beijing 100029; 3Department of Neurosurgery, Daqing Oil Field General Hospital, Daqing, Heilongjiang 163000, P.R. China Received August 12, 2016; Accepted January 4, 2018 DOI: 10.3892/mmr.2018.8552 Abstract. Allicin is an extract purified fromAllium sativum apoptotic cascades. These results implicate Allicin as a (garlic), and previous research has indicated that Allicin novel antitumor agent in treating glioma. has an inhibitory effect on many kinds of tumor cells. The aim of the present study was to explore the anticancer Introduction activity of Allicin on human glioma cells and investigate the underlying mechanism. MTT and colony‑formation Glioblastoma (GBM) is the most aggressive subset of assays were performed to detect glioma cell prolifera- primary brain tumor in adults, and is responsible for ~50% tion, and explore the effect of Allicin at various doses and of all cranial tumors. GBMs are highly infiltrative which time‑points. The apoptosis of glioma cells was measured results in difficulty for them to be resected completely (1-3). by fluorescence microscopy with Hoechst 33258 staining, Comprehensive therapy including radiotherapy and chemo- and then flow cytometry was used to analyzed changes in therapy is the main approach used for treatment; however, glioma cell apoptosis. Reverse transcription‑quantitative the overall survival of glioma patients is only 12‑14 months polymerase chain reaction and western blot analysis were post‑diagnosis (4). -

4-Hydroxynonenal, an Endogenous Aldehyde, Causes Pain and Neurogenic Inflammation Through Activation of the Irritant Receptor TRPA1
4-Hydroxynonenal, an endogenous aldehyde, causes pain and neurogenic inflammation through activation of the irritant receptor TRPA1 Marcello Trevisani*, Jan Siemens†, Serena Materazzi*, Diana M. Bautista†, Romina Nassini*, Barbara Campi‡, Noritaka Imamachi§, Eunice Andre` ‡, Riccardo Patacchini¶, Graeme S. Cottrellʈ, Raffaele Gatti‡, Allan I. Basbaum§, Nigel W. Bunnettʈ, David Julius†**, and Pierangelo Geppetti*‡** *Department of Critical Care Medicine and Surgery, Florence University, 4-50121 Florence, Italy; †Departments of Physiology and Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94143; ‡Centre of Excellence for the Study of Inflammation, University of Ferrara, 44100 Ferrara, Italy; §Departments of Anatomy and Physiology and W. M. Keck Center for Integrative Neuroscience, University of California, San Francisco, CA 94143-0444; ¶Department of Pharmacology, Chiesi Pharmaceuticals, 43100 Parma, Italy, and ʈDepartments of Surgery and Physiology, University of California, San Francisco, CA 94143 Contributed by David Julius, July 5, 2007 (sent for review June 14, 2007) TRPA1 is an excitatory ion channel expressed by a subpopulation Recent studies have shown that the wasabi receptor, TRPA1, also of primary afferent somatosensory neurons that contain sub- plays an important role in modulating nociceptor excitability and stance P and calcitonin gene-related peptide. Environmental neurogenic inflammation in the setting of tissue injury (5, 6). This irritants such as mustard oil, allicin, and acrolein activate -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Garlic and Onions: Their Cancer Prevention Properties Holly L
Published OnlineFirst January 13, 2015; DOI: 10.1158/1940-6207.CAPR-14-0172 Review Cancer Prevention Research Garlic and Onions: Their Cancer Prevention Properties Holly L. Nicastro1, Sharon A. Ross2, and John A. Milner3,† Abstract The Allium genus includes garlic, onions, shallots, leeks, and potential mechanisms of individual sulfur-containing com- chives. These vegetables are popular in cuisines worldwide and pounds and of various preparations and extracts of these are valued for their potential medicinal properties. Epidemio- vegetables, including decreased bioactivation of carcinogens, logic studies, while limited in their abilities to assess Allium antimicrobial activities, and redox modification. Allium vege- consumption, indicate some associations of Allium vegetable tables and their components have effects at each stage of consumption with decreased risk of cancer, particularly cancers carcinogenesis and affect many biologic processes that modify of the gastrointestinal tract. Limited intervention studies have cancer risk. This review discusses the cancer-preventive effects of been conducted to support these associations. The majority of Allium vegetables, particularly garlic and onions, and their supportive evidence on Allium vegetables cancer-preventive bioactive sulfur compounds and highlights research gaps. effects comes from mechanistic studies. These studies highlight Cancer Prev Res; 8(3); 181–9. Ó2015 AACR. Introduction group of foods that has raised considerable interest for their putative cancer-preventive properties is the Allium genus. Increasingly governmental entities and other organizations are Allium is the Latin word for garlic. It is part of a monocot genus proposing a wide range of food policies to promote health. These of flowering plants frequently referred to as the onion genus. -

Spice Basics
SSpicepice BasicsBasics AAllspicellspice Allspice has a pleasantly warm, fragrant aroma. The name refl ects the pungent taste, which resembles a peppery compound of cloves, cinnamon and nutmeg or mace. Good with eggplant, most fruit, pumpkins and other squashes, sweet potatoes and other root vegetables. Combines well with chili, cloves, coriander, garlic, ginger, mace, mustard, pepper, rosemary and thyme. AAnisenise The aroma and taste of the seeds are sweet, licorice like, warm, and fruity, but Indian anise can have the same fragrant, sweet, licorice notes, with mild peppery undertones. The seeds are more subtly fl avored than fennel or star anise. Good with apples, chestnuts, fi gs, fi sh and seafood, nuts, pumpkin and root vegetables. Combines well with allspice, cardamom, cinnamon, cloves, cumin, fennel, garlic, nutmeg, pepper and star anise. BBasilasil Sweet basil has a complex sweet, spicy aroma with notes of clove and anise. The fl avor is warming, peppery and clove-like with underlying mint and anise tones. Essential to pesto and pistou. Good with corn, cream cheese, eggplant, eggs, lemon, mozzarella, cheese, olives, pasta, peas, pizza, potatoes, rice, tomatoes, white beans and zucchini. Combines well with capers, chives, cilantro, garlic, marjoram, oregano, mint, parsley, rosemary and thyme. BBayay LLeafeaf Bay has a sweet, balsamic aroma with notes of nutmeg and camphor and a cooling astringency. Fresh leaves are slightly bitter, but the bitterness fades if you keep them for a day or two. Fully dried leaves have a potent fl avor and are best when dried only recently. Good with beef, chestnuts, chicken, citrus fruits, fi sh, game, lamb, lentils, rice, tomatoes, white beans. -

Chapter 4 Antimicrobial Properties of Organosulfur Compounds
Chapter 4 Antimicrobial Properties of Organosulfur Compounds Osman Sagdic and Fatih Tornuk Abstract Organosulfur compounds are defi ned as organic molecules containing one or more carbon-sulfur bonds. These compounds are present particularly in Allium and Brassica vegetables and are converted to a variety of other sulfur con- taining compounds via hydrolysis by several herbal enzymes when the intact bulbs are damaged or cut. Sulfur containing hydrolysis products constitute very diverse chemical structures and exhibit several bioactive properties as well as antimicrobial. The antimicrobial activity of organosulfur compounds has been reported against a wide spectrum of bacteria, fungi and viruses. Despite the wide antimicrobial spec- trum, their pungent fl avor/odor is the most considerable factor restricting their com- mon use in foods as antimicrobial additives. However, meat products might be considered as the most suitable food materials in this respect since Allium and Brassica vegetables especially garlic and onion have been used as fl avoring and preservative agents in meat origin foods. In this chapter, the chemical diversity and in vitro and in food antimicrobial activity of the organosulfur compounds of Allium and Brassica plants are summarized. Keywords Organosulfur compounds • Garlic • Onion • Allium • Brassica • Thiosulfi nates • Glucosinolates O. Sagdic (*) Department of Food Engineering, Faculty of Chemical and Metallurgical Engineering , Yildiz Teknik University , 34220 Esenler , Istanbul , Turkey e-mail: [email protected] F. Tornuk S a fi ye Cikrikcioglu Vocational College , Erciyes University , 38039 Kayseri , Turkey A.K. Patra (ed.), Dietary Phytochemicals and Microbes, 127 DOI 10.1007/978-94-007-3926-0_4, © Springer Science+Business Media Dordrecht 2012 128 O.