Snake Relationships Revealed by Slow-Evolving Proteins: a Preliminary Survey
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rubber Boas in Radium Hot Springs: Habitat, Inventory, and Management Strategies
Rubber Boas in Radium Hot Springs: Habitat, Inventory, and Management Strategies ROBERT C. ST. CLAIR1 AND ALAN DIBB2 19809 92 Avenue, Edmonton, AB, T6E 2V4, Canada, email [email protected] 2Parks Canada, Box 220, Radium Hot Springs, BC, V0A 1M0, Canada Abstract: Radium Hot Springs in Kootenay National Park, British Columbia is home to a population of rubber boas (Charina bottae). This population is of ecological and physiological interest because the hot springs seem to be a thermal resource for the boas, which are near the northern limit of their range. This population also presents a dilemma to park management because the site is a major tourist destination and provides habitat for the rubber boa, which is listed as a species of Special Concern by the Committee on the Status of Endangered Wildlife in Canada (COSEWIC). An additional dilemma is that restoration projects, such as prescribed logging and burning, may increase available habitat for the species but kill individual snakes. Successful management of the population depends on monitoring the population, assessing the impacts of restoration strategies, and mapping both summer and winter habitat. Hibernation sites may be discovered only by using radiotelemetry to follow individuals. Key Words: rubber boa, Charina bottae, Radium Hot Springs, hot springs, snakes, habitat, restoration, Kootenay National Park, British Columbia Introduction The presence of rubber boas (Charina bottae) at Radium Hot Springs in Kootenay National Park, British Columbia (B.C.) presents some unusual challenges for wildlife managers because the site is a popular tourist resort and provides habitat for the species, which is listed as a species of Special Concern by the Committee on the Status of Endangered Wildlife in Canada (COSEWIC 2004). -
Reptiles (Viviparous Lizards and Slow-Worms Anguis REFERENCES Fragilis)
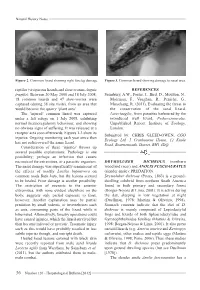
Natural History Notes Figure 2. Common lizard showing right foreleg damage. Figure 3. Common lizard showing damage to nasal area. reptiles (viviparous lizards and slow-worms Anguis REFERENCES fragilis). Between 20 May 2008 and 18 July 2008, Sainsbury, A.W., Foster, J., Bird, D., Moulton, N., 18 common lizards and 47 slow-worms were Molenaar, F., Vaughan, R., Peniche, G., captured (during 38 site visits), from an area that Marschang, R. (2011). Evaluating the threat to would become the quarry ‘plant area’. the conservation of the sand lizard, The ‘injured’ common lizard was captured Lacertaagilis, from parasites harboured by the under a felt refuge on 1 July 2008, exhibiting introduced wall lizard, Podarcismuralis. normal thermoregulatory behaviour, and showing Unpublished Report. Institute of Zoology, no obvious signs of suffering. It was released at a London. receptor area soon afterwards. Figures 1-3 show its Submitted by: CHRIS GLEED-OWEN, CGO injuries. Ongoing monitoring each year since then Ecology Ltd, 5 Cranbourne House, 12 Knole has not rediscovered the same lizard. Road, Bournemouth, Dorset, BH1 4DQ. Consideration of these ‘injuries’ throws up several possible explanations. Pathology is one possibility; perhaps an infection that causes necrosis of the extremities, or a parasitic organism. DRYMOLUBER DICHROUS (northern The nasal damage was superficially reminiscent of woodland racer) and ANOLIS FUSCOAURATUS the effects of toadfly Lucilia bufonivora on (slender anole): PREDATION. common toads Bufo bufo, but the lesions seemed Drymoluber dichrous (Peters, 1863) is a ground- to be healed. Frost damage is another possibility. dwelling colubrid from northern South America The restriction of necrosis to the anterior found in both primary and secondary forest extremities, with none evident elsewhere on the (Borges-Nojosa & Lima, 2001). -

ONEP V09.Pdf
Compiled by Jarujin Nabhitabhata Tanya Chan-ard Yodchaiy Chuaynkern OEPP BIODIVERSITY SERIES volume nine OFFICE OF ENVIRONMENTAL POLICY AND PLANNING MINISTRY OF SCIENCE TECHNOLOGY AND ENVIRONMENT 60/1 SOI PIBULWATTANA VII, RAMA VI RD., BANGKOK 10400 THAILAND TEL. (662) 2797180, 2714232, 2797186-9 FAX. (662) 2713226 Office of Environmental Policy and Planning 2000 NOT FOR SALE NOT FOR SALE NOT FOR SALE Compiled by Jarujin Nabhitabhata Tanya Chan-ard Yodchaiy Chuaynkern Office of Environmental Policy and Planning 2000 First published : September 2000 by Office of Environmental Policy and Planning (OEPP), Thailand. ISBN : 974–87704–3–5 This publication is financially supported by OEPP and may be reproduced in whole or in part and in any form for educational or non–profit purposes without special permission from OEPP, providing that acknowledgment of the source is made. No use of this publication may be made for resale or for any other commercial purposes. Citation : Nabhitabhata J., Chan ard T., Chuaynkern Y. 2000. Checklist of Amphibians and Reptiles in Thailand. Office of Environmental Policy and Planning, Bangkok, Thailand. Authors : Jarujin Nabhitabhata Tanya Chan–ard Yodchaiy Chuaynkern National Science Museum Available from : Biological Resources Section Natural Resources and Environmental Management Division Office of Environmental Policy and Planning Ministry of Science Technology and Environment 60/1 Rama VI Rd. Bangkok 10400 THAILAND Tel. (662) 271–3251, 279–7180, 271–4232–8 279–7186–9 ext 226, 227 Facsimile (662) 279–8088, 271–3251 Designed & Printed :Integrated Promotion Technology Co., Ltd. Tel. (662) 585–2076, 586–0837, 913–7761–2 Facsimile (662) 913–7763 2 1. -

De Los Reptiles Del Yasuní
guía dinámica de los reptiles del yasuní omar torres coordinador editorial Lista de especies Número de especies: 113 Amphisbaenia Amphisbaenidae Amphisbaena bassleri, Culebras ciegas Squamata: Serpentes Boidae Boa constrictor, Boas matacaballo Corallus hortulanus, Boas de los jardines Epicrates cenchria, Boas arcoiris Eunectes murinus, Anacondas Colubridae: Dipsadinae Atractus major, Culebras tierreras cafés Atractus collaris, Culebras tierreras de collares Atractus elaps, Falsas corales tierreras Atractus occipitoalbus, Culebras tierreras grises Atractus snethlageae, Culebras tierreras Clelia clelia, Chontas Dipsas catesbyi, Culebras caracoleras de Catesby Dipsas indica, Culebras caracoleras neotropicales Drepanoides anomalus, Culebras hoz Erythrolamprus reginae, Culebras terrestres reales Erythrolamprus typhlus, Culebras terrestres ciegas Erythrolamprus guentheri, Falsas corales de nuca rosa Helicops angulatus, Culebras de agua anguladas Helicops pastazae, Culebras de agua de Pastaza Helicops leopardinus, Culebras de agua leopardo Helicops petersi, Culebras de agua de Peters Hydrops triangularis, Culebras de agua triángulo Hydrops martii, Culebras de agua amazónicas Imantodes lentiferus, Cordoncillos del Amazonas Imantodes cenchoa, Cordoncillos comunes Leptodeira annulata, Serpientes ojos de gato anilladas Oxyrhopus petolarius, Falsas corales amazónicas Oxyrhopus melanogenys, Falsas corales oscuras Oxyrhopus vanidicus, Falsas corales Philodryas argentea, Serpientes liana verdes de banda plateada Philodryas viridissima, Serpientes corredoras -

Zootaxa, Molecular Phylogeny, Classification, and Biogeography Of
Zootaxa 2067: 1–28 (2009) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2009 · Magnolia Press ISSN 1175-5334 (online edition) Molecular phylogeny, classification, and biogeography of West Indian racer snakes of the Tribe Alsophiini (Squamata, Dipsadidae, Xenodontinae) S. BLAIR HEDGES1, ARNAUD COULOUX2, & NICOLAS VIDAL3,4 1Department of Biology, 208 Mueller Lab, Pennsylvania State University, University Park, PA 16802-5301 USA. E-mail: [email protected] 2Genoscope. Centre National de Séquençage, 2 rue Gaston Crémieux, CP5706, 91057 Evry Cedex, France www.genoscope.fr 3UMR 7138, Département Systématique et Evolution, Muséum National d’Histoire Naturelle, CP 26, 57 rue Cuvier, 75005 Paris, France 4Corresponding author. E-mail : [email protected] Abstract Most West Indian snakes of the family Dipsadidae belong to the Subfamily Xenodontinae and Tribe Alsophiini. As recognized here, alsophiine snakes are exclusively West Indian and comprise 43 species distributed throughout the region. These snakes are slender and typically fast-moving (active foraging), diurnal species often called racers. For the last four decades, their classification into six genera was based on a study utilizing hemipenial and external morphology and which concluded that their biogeographic history involved multiple colonizations from the mainland. Although subsequent studies have mostly disagreed with that phylogeny and taxonomy, no major changes in the classification have been proposed until now. Here we present a DNA sequence analysis of five mitochondrial genes and one nuclear gene in 35 species and subspecies of alsophiines. Our results are more consistent with geography than previous classifications based on morphology, and support a reclassification of the species of alsophiines into seven named and three new genera: Alsophis Fitzinger (Lesser Antilles), Arrhyton Günther (Cuba), Borikenophis Hedges & Vidal gen. -

Review of Species Selected at SRG 45 Following Working Group Recommendations on Reptiles and One Scorpion from Benin, Ghana and Togo
Review of species selected at SRG 45 following working group recommendations on reptiles and one scorpion from Benin, Ghana and Togo (Version edited for public release) A report to the European Commission Directorate General E - Environment ENV.E.2. – Environmental Agreements and Trade by the United Nations Environment Programme World Conservation Monitoring Centre November, 2008 UNEP World Conservation Monitoring Centre 219 Huntingdon Road Cambridge CB3 0DL CITATION United Kingdom UNEP-WCMC (2008). Review of species selected at SRG 45 following working group recommendations on reptiles Tel: +44 (0) 1223 277314 and one scorpion from Benin, Ghana and Togo. A Report Fax: +44 (0) 1223 277136 to the European Commission. UNEP-WCMC, Cambridge. Email: [email protected] Website: www.unep-wcmc.org PREPARED FOR ABOUT UNEP-WORLD CONSERVATION The European Commission, Brussels, Belgium MONITORING CENTRE The UNEP World Conservation Monitoring Centre DISCLAIMER (UNEP-WCMC), based in Cambridge, UK, is the The contents of this report do not necessarily reflect specialist biodiversity information and assessment the views or policies of UNEP or contributory centre of the United Nations Environment organisations. The designations employed and the Programme (UNEP), run cooperatively with WCMC presentations do not imply the expressions of any 2000, a UK charity. The Centre's mission is to opinion whatsoever on the part of UNEP, the evaluate and highlight the many values of European Commission or contributory organisations biodiversity and put authoritative biodiversity concerning the legal status of any country, territory, knowledge at the centre of decision-making. city or area or its authority, or concerning the Through the analysis and synthesis of global delimitation of its frontiers or boundaries. -

New Records of Snakes (Squamata: Serpentes) from Hoa Binh Province, Northwestern Vietnam
Bonn zoological Bulletin 67 (1): 15–24 May 2018 New records of snakes (Squamata: Serpentes) from Hoa Binh Province, northwestern Vietnam Truong Quang Nguyen1,2,*, Tan Van Nguyen 1,3, Cuong The Pham1,2, An Vinh Ong4 & Thomas Ziegler5 1 Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 2 Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 3 Save Vietnam’s Wildlife, Cuc Phuong National Park, Ninh Binh Province, Vietnam 4 Vinh University, 182 Le Duan Road, Vinh City, Nghe An Province, Vietnam 5 AG Zoologischer Garten Köln, Riehler Strasse 173, D-50735 Cologne, Germany * Corresponding author. E-mail: [email protected] Abstract. We report nine new records of snakes from Hoa Binh Province based on a reptile collection from Thuong Tien, Hang Kia-Pa Co, Ngoc Son-Ngo Luong nature reserves, and Tan Lac District, comprising six species of Colubri- dae (Dryocalamus davisonii, Euprepiophis mandarinus, Lycodon futsingensis, L. meridionalis, Sibynophis collaris and Sinonatrix aequifasciata), one species of Pareatidae (Pareas hamptoni) and two species of Viperidae (Protobothrops mu- crosquamatus and Trimeresurus gumprechti). In addition, we provide an updated list of 43 snake species from Hoa Binh Province. The snake fauna of Hoa Binh contains some species of conservation concern with seven species listed in the Governmental Decree No. 32/2006/ND-CP (2006), nine species listed in the Vietnam Red Data Book (2007), and three species listed in the IUCN Red List (2018). Key words. New records, snakes, taxonomy, Hoa Binh Province. -

Genus Lycodon)
Zoologica Scripta Multilocus phylogeny reveals unexpected diversification patterns in Asian wolf snakes (genus Lycodon) CAMERON D. SILER,CARL H. OLIVEROS,ANSSI SANTANEN &RAFE M. BROWN Submitted: 6 September 2012 Siler, C. D., Oliveros, C. H., Santanen, A., Brown, R. M. (2013). Multilocus phylogeny Accepted: 8 December 2012 reveals unexpected diversification patterns in Asian wolf snakes (genus Lycodon). —Zoologica doi:10.1111/zsc.12007 Scripta, 42, 262–277. The diverse group of Asian wolf snakes of the genus Lycodon represents one of many poorly understood radiations of advanced snakes in the superfamily Colubroidea. Outside of three species having previously been represented in higher-level phylogenetic analyses, nothing is known of the relationships among species in this unique, moderately diverse, group. The genus occurs widely from central to Southeast Asia, and contains both widespread species to forms that are endemic to small islands. One-third of the diversity is found in the Philippine archipelago. Both morphological similarity and highly variable diagnostic characters have contributed to confusion over species-level diversity. Additionally, the placement of the genus among genera in the subfamily Colubrinae remains uncertain, although previous studies have supported a close relationship with the genus Dinodon. In this study, we provide the first estimate of phylogenetic relationships within the genus Lycodon using a new multi- locus data set. We provide statistical tests of monophyly based on biogeographic, morpho- logical and taxonomic hypotheses. With few exceptions, we are able to reject many of these hypotheses, indicating a need for taxonomic revisions and a reconsideration of the group's biogeography. Mapping of color patterns on our preferred phylogenetic tree suggests that banded and blotched types have evolved on multiple occasions in the history of the genus, whereas the solid-color (and possibly speckled) morphotype color patterns evolved only once. -

Snakes of South-East Asia Including Myanmar, Thailand, Malaysia, Singapore, Sumatra, Borneo, Java and Bali
A Naturalist’s Guide to the SNAKES OF SOUTH-EAST ASIA including Myanmar, Thailand, Malaysia, Singapore, Sumatra, Borneo, Java and Bali Indraneil Das First published in the United Kingdom in 2012 by Beaufoy Books n n 11 Blenheim Court, 316 Woodstock Road, Oxford OX2 7NS, England Contents www.johnbeaufoy.com 10 9 8 7 6 5 4 3 2 1 Introduction 4 Copyright © 2012 John Beaufoy Publishing Limited Copyright in text © Indraneil Das Snake Topography 4 Copyright in photographs © [to come] Dealing with Snake Bites 6 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without the prior written permission of the publishers. About this Book 7 ISBN [to come] Glossary 8 Edited, designed and typeset by D & N Publishing, Baydon, Wiltshire, UK Printed and bound [to come] Species Accounts and Photographs 11 Checklist of South-East Asian Snakes 141 Dedication Nothing would have happened without the support of the folks at home: my wife, Genevieve V.A. Gee, and son, Rahul Das. To them, I dedicate this book. Further Reading 154 Acknowledgements 155 Index 157 Edited and designed by D & N Publishing, Baydon, Wiltshire, UK Printed and bound in Malaysia by Times Offset (M) Sdn. Bhd. n Introduction n n Snake Topography n INTRODUCTION Snakes form one of the major components of vertebrate fauna of South-East Asia. They feature prominently in folklore, mythology and other belief systems of the indigenous people of the region, and are of ecological and conservation value, some species supporting significant (albeit often illegal) economic activities (primarily, the snake-skin trade, but also sale of meat and other body parts that purportedly have medicinal properties). -

The Reptile Collection of the Museu De Zoologia, Pecies
Check List 9(2): 257–262, 2013 © 2013 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution The Reptile Collection of the Museu de Zoologia, PECIES S Universidade Federal da Bahia, Brazil OF Breno Hamdan 1,2*, Daniela Pinto Coelho 1 1, Eduardo José dos Reis Dias3 ISTS 1 L and Rejâne Maria Lira-da-Silva , Annelise Batista D’Angiolella 40170-115, Salvador, BA, Brazil. 1 Universidade Federal da Bahia, Instituto de Biologia, Departamento de Zoologia, Núcleo Regional de Ofiologia e Animais Peçonhentos. CEP Sala A0-92 (subsolo), Laboratório de Répteis, Ilha do Fundão, Av. Carlos Chagas Filho, N° 373. CEP 21941-902. Rio de Janeiro, RJ, Brazil. 2 Programa de Pós-Graduação em Zoologia, Museu Nacional/UFRJ. Universidade Federal do Rio de Janeiro Centro de Ciências da Saúde, Bloco A, Carvalho. CEP 49500-000. Itabaian, SE, Brazil. * 3 CorrUniversidadeesponding Federal author. de E-mail: Sergipe, [email protected] Departamento de Biociências, Laboratório de Biologia e Ecologia de Vertebrados (LABEV), Campus Alberto de Abstract: to its history. The Reptile Collection of the Museu de Zoologia from Universidade Federal da Bahia (CRMZUFBA) has 5,206 specimens and Brazilian 185 species scientific (13 collections endemic to represent Brazil and an 9important threatened) sample with of one the quarter country’s of biodiversitythe known reptile and are species a testament listed in Brazil, from over 175 municipalities. Although the CRMZUFBA houses species from all Brazilian biomes there is a strong regional presence. Knowledge of the species housed in smaller collections could avoid unrepresentative species descriptions and provide information concerning intraspecific variation, ecological features and geographic coverage. -

Xenosaurus Tzacualtipantecus. the Zacualtipán Knob-Scaled Lizard Is Endemic to the Sierra Madre Oriental of Eastern Mexico
Xenosaurus tzacualtipantecus. The Zacualtipán knob-scaled lizard is endemic to the Sierra Madre Oriental of eastern Mexico. This medium-large lizard (female holotype measures 188 mm in total length) is known only from the vicinity of the type locality in eastern Hidalgo, at an elevation of 1,900 m in pine-oak forest, and a nearby locality at 2,000 m in northern Veracruz (Woolrich- Piña and Smith 2012). Xenosaurus tzacualtipantecus is thought to belong to the northern clade of the genus, which also contains X. newmanorum and X. platyceps (Bhullar 2011). As with its congeners, X. tzacualtipantecus is an inhabitant of crevices in limestone rocks. This species consumes beetles and lepidopteran larvae and gives birth to living young. The habitat of this lizard in the vicinity of the type locality is being deforested, and people in nearby towns have created an open garbage dump in this area. We determined its EVS as 17, in the middle of the high vulnerability category (see text for explanation), and its status by the IUCN and SEMAR- NAT presently are undetermined. This newly described endemic species is one of nine known species in the monogeneric family Xenosauridae, which is endemic to northern Mesoamerica (Mexico from Tamaulipas to Chiapas and into the montane portions of Alta Verapaz, Guatemala). All but one of these nine species is endemic to Mexico. Photo by Christian Berriozabal-Islas. Amphib. Reptile Conserv. | http://redlist-ARC.org 01 June 2013 | Volume 7 | Number 1 | e61 Copyright: © 2013 Wilson et al. This is an open-access article distributed under the terms of the Creative Com- mons Attribution–NonCommercial–NoDerivs 3.0 Unported License, which permits unrestricted use for non-com- Amphibian & Reptile Conservation 7(1): 1–47. -

Download Download
HAMADRYAD Vol. 27. No. 2. August, 2003 Date of issue: 31 August, 2003 ISSN 0972-205X CONTENTS T. -M. LEONG,L.L.GRISMER &MUMPUNI. Preliminary checklists of the herpetofauna of the Anambas and Natuna Islands (South China Sea) ..................................................165–174 T.-M. LEONG & C-F. LIM. The tadpole of Rana miopus Boulenger, 1918 from Peninsular Malaysia ...............175–178 N. D. RATHNAYAKE,N.D.HERATH,K.K.HEWAMATHES &S.JAYALATH. The thermal behaviour, diurnal activity pattern and body temperature of Varanus salvator in central Sri Lanka .........................179–184 B. TRIPATHY,B.PANDAV &R.C.PANIGRAHY. Hatching success and orientation in Lepidochelys olivacea (Eschscholtz, 1829) at Rushikulya Rookery, Orissa, India ......................................185–192 L. QUYET &T.ZIEGLER. First record of the Chinese crocodile lizard from outside of China: report on a population of Shinisaurus crocodilurus Ahl, 1930 from north-eastern Vietnam ..................193–199 O. S. G. PAUWELS,V.MAMONEKENE,P.DUMONT,W.R.BRANCH,M.BURGER &S.LAVOUÉ. Diet records for Crocodylus cataphractus (Reptilia: Crocodylidae) at Lake Divangui, Ogooué-Maritime Province, south-western Gabon......................................................200–204 A. M. BAUER. On the status of the name Oligodon taeniolatus (Jerdon, 1853) and its long-ignored senior synonym and secondary homonym, Oligodon taeniolatus (Daudin, 1803) ........................205–213 W. P. MCCORD,O.S.G.PAUWELS,R.BOUR,F.CHÉROT,J.IVERSON,P.C.H.PRITCHARD,K.THIRAKHUPT, W. KITIMASAK &T.BUNDHITWONGRUT. Chitra burmanica sensu Jaruthanin, 2002 (Testudines: Trionychidae): an unavailable name ............................................................214–216 V. GIRI,A.M.BAUER &N.CHATURVEDI. Notes on the distribution, natural history and variation of Hemidactylus giganteus Stoliczka, 1871 ................................................217–221 V. WALLACH.