Resistant Staphylococcus Aureus Clinical Strains from The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Erythromycin Versus Tetracycline for Treatment of Mediterranean Spotted Fever
Arch Dis Child: first published as 10.1136/adc.61.10.1027 on 1 October 1986. Downloaded from Archives of Disease in Childhood, 1986, 61, 1027-1029 Erythromycin versus tetracycline for treatment of Mediterranean spotted fever T MUNOZ-ESPIN, P LOPEZ-PARtS, E ESPEJO-ARENAS, B FONT-CREUS, I MARTINEZ- VILA, J TRAVERIA-CASANOVA, F SEGURA-PORTA, AND F BELLA-CUETO Hospital de Sant Llatzer, Terrassa, Clinica Infantil del Nen Jesus, Sabadell, and Hospital Mare de Deu de la Salut, Sabadell, Barcelona, Spain SUMMARY Eighty one children aged between 1 and 13 years participated in a randomised comparative trial of tetracycline hydrochloride and erythromycin stearate for treatment of Mediterranean spotted fever. Both therapeutic regimens proved effective, but in patients treated with tetracycline both clinical symptoms and fever disappeared significantly more quickly. Likewise, when those patients who began treatment within the first 72 hours of illness are considered the febrile period had a significantly shorter duration in the group treated with tetracycline. One patient was switched to tetracycline because there was no improvement of clinical manifestations, with persistence of fever, myalgias, and prostration, after receiving eight days of treatment with erythromycin. These results suggest that tetracyclines are superior to erythromycin in the treatment of Mediterranean spotted fever. copyright. Mediterranean spotted fever is an acute infectious were not included in the trial; neither were those disease caused by Rickettsia conorii. During the -

Use of Antibiotics to Prevent Calf Diarrhea and Septiceinia
PEER REVIEWED Use of Antibiotics to Prevent Calf Diarrhea and Septiceinia Peter D. Constable, BVSc, PhD, DACVIM Department of Veterinary Clinical Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61802 Abstract l'oxytetracycline (0.15 to 6.0 mg/lb [0.32 to 13.2 mg/kg] au 24 h, per os) sont efficaces dans le traitement de la This article reviews studies related to the use of diarrhee chez les veaux et permettent un accroissement antimicrobial agents to prevent calf diarrhea and septi du taux de croissance des veaux nourris au lait. De cemia, and discusses whether prophylactic administra plus, la chlortetracycline permet la diminution du taux tion of antibiotics in neonatal calves is effective and de mortalite lorsque administree chez les veaux indicated. Orally administered chlortetracycline, oxytet nouveau-nes pour prevenir la diarrhee. 11 n'y a pas racycline, tetracycline and neomycin have label claims d'antibiotiques qui promettent de prevenir la in the US for the "control" or "aid in the control" of calf septicemie chez les veaux. Paree qu'il ne semble pas diarrhea caused by bacteria susceptible to the antibi exister d'etudes sur l'efficacite de la tetracycline et de otic. Chlortetracycline and oxytetracycline (0.15 to 6.0 la neomycine et parce que !'utilisation hors homologa mg/lb [0.32 to 13.2 mg/kg], q 24 h, PO) are efficacious tion des medicaments. n'est pas permise pour la for preventing calf diarrhea and increasing growth rate prevention routiniere des maladies ou !'augmentation in milk-fed calves, and chlortetracycline (3 mg/lb [7 mg/ du taux de croissance et du taux de conversion kg], q 12 h, PO) is efficacious for decreasing mortality alimentaire, selon le code de l'AMDUCA de 1994, seule when administered to prevent diarrhea in neonatal !'administration orale de chlortetracycline et calves. -

Long-Term Use of Spironolactone for Acne in Women: a Case Series of 403 Patients
Long-term use of spironolactone for acne in women: A case series of 403 patients Vaibhav Garg, BS,a JulianaK.Choi,MD,PhD,b William D. James, MD,b and John S. Barbieri, MD, MBAb Philadelphia, Pennsylvania Background: There are limited data regarding the long-term outcomes of spironolactone use for women with acne and its effect on truncal acne. Objective: To comprehensively describe outcomes of patients treated with spironolactone in routine clinical practice, including long-term outcomes. Methods: We performed a retrospective case series of 403 adult women treated for acne with spironolactone at an academic medical center between 2008 and 2019. Rates of objective, as assessed by Comprehensive Acne Severity Scale scores, and subjective acne clearance were evaluated, as well as rates of treatment discontinuation, dosage changes, and drug survival. Logistic regression was used to assess for association between incidence of menstrual adverse effects and combined oral contraceptive use. Results: As evaluated by Comprehensive Acne Severity Scale scores, at the first follow-up, 75.5%, 84.0%, and 80.2% of patients with available data had reduction or complete clearance of acne on the face, chest, and back, respectively. The mean drug survival was 470.7 days. Menstrual adverse effects were less common among those using combined oral contraception (odds ratio, 0.23; 95% confidence interval, 0.11-0.50). Limitations: This study was conducted at a single academic medical center. Conclusions: Spironolactone improves clinical outcomes and is well tolerated for many adult women with acne using it for an extended duration. ( J Am Acad Dermatol 2021;84:1348-55.) Key words: acne; acne vulgaris; birth control pill; combined oral contraceptive; comprehensive acne severity scale; oral antibiotics; outcomes; spironolactone. -

Prednisolone / Neomycin / Tetracycline For- Mulation
SAFETY DATA SHEET Prednisolone / Neomycin / Tetracycline For- mulation Version Revision Date: SDS Number: Date of last issue: 25.08.2020 3.6 09.04.2021 407521-00015 Date of first issue: 07.01.2016 SECTION 1: Identification of the substance/mixture and of the company/undertaking 1.1 Product identifier Trade name : Prednisolone / Neomycin / Tetracycline Formulation 1.2 Relevant identified uses of the substance or mixture and uses advised against Use of the Sub- : Veterinary product stance/Mixture 1.3 Details of the supplier of the safety data sheet Company : MSD 20 Spartan Road 1619 Spartan, South Africa Telephone : +27119239300 E-mail address of person : [email protected] responsible for the SDS 1.4 Emergency telephone number +1-908-423-6000 SECTION 2: Hazards identification 2.1 Classification of the substance or mixture Classification (REGULATION (EC) No 1272/2008) Skin sensitisation, Category 1 H317: May cause an allergic skin reaction. Reproductive toxicity, Category 1A H360D: May damage the unborn child. Effects on or via lactation H362: May cause harm to breast-fed children. Short-term (acute) aquatic hazard, Cate- H400: Very toxic to aquatic life. gory 1 Long-term (chronic) aquatic hazard, Cat- H410: Very toxic to aquatic life with long lasting egory 1 effects. 2.2 Label elements Labelling (REGULATION (EC) No 1272/2008) Hazard pictograms : Signal word : Danger Hazard statements : H317 May cause an allergic skin reaction. H360D May damage the unborn child. H362 May cause harm to breast-fed children. H410 Very toxic to aquatic life with long lasting effects. 1 / 23 SAFETY DATA SHEET Prednisolone / Neomycin / Tetracycline For- mulation Version Revision Date: SDS Number: Date of last issue: 25.08.2020 3.6 09.04.2021 407521-00015 Date of first issue: 07.01.2016 Precautionary statements : Prevention: P201 Obtain special instructions before use. -
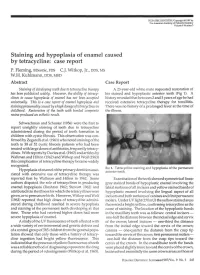
Staining and Hypoplasia of Enamel Caused by Tetracycline: Case Report P
PEDIATRIC DENTISTRY/Copyright © 1987 by The American Academy of Pediatric Dentistry Volume 9 Number 3 Staining and hypoplasia of enamel caused by tetracycline: case report P. Fleming, BDentSc, FDS C.J. Witkop, Jr., DDS, MS W.H. Kuhlmann, DDS, MSD Abstract Case Report Staining of developing teeth due to tetracycline therapy A 23-year-old white male requested restoration of has been publicized widely. However, the ability of tetracy- his stained and hypoplastic anterior teeth (Fig 1). A clines to cause hypoplasia of enamel has not been accepted history revealed that between 2 and 3 years of age he had universally. This is a case report of enamel hypoplasia and received extensive tetracycline therapy for tonsillitis. staining presumably caused byahigh dosage of'tetracy'dines in There was no history of a prolonged fever at the time of childhood. Restoration of the teeth with bonded composite the illness. resins produced an esthetic result. -UK Schwachman and Schuster (1956) were the first to report unsightly staining of teeth due to tetracycline administered during the period of tooth formation in children with cystic fibrosis. This observation was con- y firmed by Zegarelli et al. (1961) who noted staining of the teeth in 38 of 52 cystic fibrosis patients who had been ijgj treated with large doses of antibiotics, frequently tetracy- .^••W^^ clines. With reports by Davies et al. (1962) and articles by Wallman and Hilton (1962) and Witkop and Wolf (1963) this complication of tetracycline therapy became widely recognized. FIG 1. Tetracycline staining and hypoplasia of the permanent Hypoplasia of enamel of the primary dentition asso- anterior teeth. -

Prednisolone / Neomycin / Tetracycline For- Mulation
SAFETY DATA SHEET Prednisolone / Neomycin / Tetracycline For- mulation Version Revision Date: SDS Number: Date of last issue: 23.03.2020 5.5 25.08.2020 407516-00013 Date of first issue: 07.01.2016 SECTION 1. PRODUCT AND COMPANY IDENTIFICATION Product name : Prednisolone / Neomycin / Tetracycline Formulation Manufacturer or supplier's details Company name of supplier : MSD Address : 2000 Galloping Hill Road Kenilworth - New Jersey - U.S.A. 07033 Telephone : 908-740-4000 Emergency telephone : 1-908-423-6000 E-mail address : [email protected] Recommended use of the chemical and restrictions on use Recommended use : Veterinary product SECTION 2. HAZARDS IDENTIFICATION GHS Classification Skin sensitization : Category 1 Reproductive toxicity : Category 1A Effects on or via lactation Specific target organ toxicity : Category 2 (Kidney, inner ear) - repeated exposure Specific target organ toxicity : Category 2 (Gastrointestinal tract, Nervous system, Skin, Teeth) - repeated exposure (Oral) GHS label elements Hazard pictograms : Signal Word : Danger Hazard Statements : H317 May cause an allergic skin reaction. H360D May damage the unborn child. H362 May cause harm to breast-fed children. H373 May cause damage to organs (Kidney, inner ear) through prolonged or repeated exposure. H373 May cause damage to organs (Gastrointestinal tract, Nervous system, Skin, Teeth) through prolonged or repeated exposure if swallowed. Precautionary Statements : Prevention: P201 Obtain special instructions before use. P202 Do not handle until all safety precautions have been read and understood. 1 / 24 SAFETY DATA SHEET Prednisolone / Neomycin / Tetracycline For- mulation Version Revision Date: SDS Number: Date of last issue: 23.03.2020 5.5 25.08.2020 407516-00013 Date of first issue: 07.01.2016 P260 Do not breathe dust. -

2021 Formulary List of Covered Prescription Drugs
2021 Formulary List of covered prescription drugs This drug list applies to all Individual HMO products and the following Small Group HMO products: Sharp Platinum 90 Performance HMO, Sharp Platinum 90 Performance HMO AI-AN, Sharp Platinum 90 Premier HMO, Sharp Platinum 90 Premier HMO AI-AN, Sharp Gold 80 Performance HMO, Sharp Gold 80 Performance HMO AI-AN, Sharp Gold 80 Premier HMO, Sharp Gold 80 Premier HMO AI-AN, Sharp Silver 70 Performance HMO, Sharp Silver 70 Performance HMO AI-AN, Sharp Silver 70 Premier HMO, Sharp Silver 70 Premier HMO AI-AN, Sharp Silver 73 Performance HMO, Sharp Silver 73 Premier HMO, Sharp Silver 87 Performance HMO, Sharp Silver 87 Premier HMO, Sharp Silver 94 Performance HMO, Sharp Silver 94 Premier HMO, Sharp Bronze 60 Performance HMO, Sharp Bronze 60 Performance HMO AI-AN, Sharp Bronze 60 Premier HDHP HMO, Sharp Bronze 60 Premier HDHP HMO AI-AN, Sharp Minimum Coverage Performance HMO, Sharp $0 Cost Share Performance HMO AI-AN, Sharp $0 Cost Share Premier HMO AI-AN, Sharp Silver 70 Off Exchange Performance HMO, Sharp Silver 70 Off Exchange Premier HMO, Sharp Performance Platinum 90 HMO 0/15 + Child Dental, Sharp Premier Platinum 90 HMO 0/20 + Child Dental, Sharp Performance Gold 80 HMO 350 /25 + Child Dental, Sharp Premier Gold 80 HMO 250/35 + Child Dental, Sharp Performance Silver 70 HMO 2250/50 + Child Dental, Sharp Premier Silver 70 HMO 2250/55 + Child Dental, Sharp Premier Silver 70 HDHP HMO 2500/20% + Child Dental, Sharp Performance Bronze 60 HMO 6300/65 + Child Dental, Sharp Premier Bronze 60 HDHP HMO -

Spironolactone Information Sheet
Spironolactone Information Sheet Spironolactone has been used off-license in women with acne for over 30 years due to its anti-androgenic properties and is included in American Guidelines for acne. However, although widely used by some dermatologists in the UK, there is little convincing trial evidence of its benefit. What dose of spironolactone is prescribed for adult female acne? SAFA participants are prescribed spironolactone (or placebo) 50mg once daily for 6 weeks, increased to 100mg once daily for a further 18 weeks if tolerated. Effectiveness of treatment is judged after 12 weeks. The reason for dose escalation is to avoid side effects from spironolactone, which can cause gastro-intestinal disturbances, dizziness, gynaecomastia and menstrual disturbances, particularly at higher doses. Is potassium and renal monitoring required? 1 American guidelines and large UK-based observational datasets suggest that, providing a baseline renal function test is normal then no further monitoring is required. All participants in the SAFA trial have renal function tests, including potassium, carried out at baseline, so do not need further blood testing. Is spironolactone teratogenic? Spironolactone should be avoided in pregnancy but risk of harm to the foetus is thought to be less high than 1,2 for other oral acne treatments, such as tetracycline antibiotics, co-cyprindiol and oral isotretinoin. Pregnancy risk relates to feminisation of the male foetus in the last trimester. Pregnancy tests are carried out at baseline in the SAFA trial and participants of child-bearing potential at risk of pregnancy are advised they must use effective contraception. Why isn’t spironolactone used for men with acne? A small study found that a many young men with acne who were treated with spironolactone experienced breast enlargement and the trial was stopped early. -
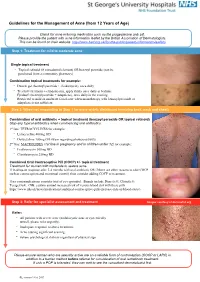
Guidelines for the Management of Acne (From 12 Years of Age)
Guidelines for the Management of Acne (from 12 Years of Age) Check for acne inducing medication such as the progesterone only pill. Please provide the patient with acne information leaflet by the British Association of Dermatologists. This can be found on their website: http://www.bad.org.uk/for-the-public/patient-information-leaflets Step 1: Treatment for mild to moderate acne Single topical treatment - Topical retinoid (if comedomal element) OR benzoyl peroxide (can be purchased from a community pharmacy) Combination topical treatments for example: - Duac® gel (benzoyl peroxide + clindamycin), once daily - Treclin® (tretinoin + clindamycin), apply thinly once daily at bedtime Epiduo® (benzoylperoxide + adapalene), once daily in the evening. Restricted to mild or moderate facial acne when monotherapy with benzoyl peroxide or adapalene is not sufficient. StepStep 2:2: WhenWhen notnot respondingresponding toto StepStep 11 (orand more 2 (or widely more widelydistributed distributed involving involving back, neckback, and neck chest) and chest) Combination of oral antibiotic + topical treatment (benzoyl peroxide OR topical retinoid) Stop any topical antibiotics when commencing oral antibiotics 1st line: TETRACYCLINES for example: - Lymecycline 408mg OD - Doxycycline 100mg OD (warn regarding photosensitivity) 2nd line: MACROLIDES (1st line in pregnancy and in children under 12) for example: - Erythromycin 500mg BD - Clarithromycin 250mg BD Combined Oral Contraceptive Pill (COCP) +/- topical treatment Treatment for women with moderate to severe acne. If inadequate response after 3-4 months with oral antibiotic OR if there are other reasons to take COCP such as contraception and menstrual control, then consider adding COCP to treatment. If no contraindications consider trial of co-cyprindiol. Brands include Dianette®, Clairette®, Teragezza®. -

Lactams and Florfenicol Antibiotics Remain Bioactive in Soils While Ciprofloxacin, Neomycin, and Tetracycline Are Neutralizedᰔ Murugan Subbiah,1 Shannon M
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Oct. 2011, p. 7255–7260 Vol. 77, No. 20 0099-2240/11/$12.00 doi:10.1128/AEM.05352-11 Copyright © 2011, American Society for Microbiology. All Rights Reserved. -Lactams and Florfenicol Antibiotics Remain Bioactive in Soils while Ciprofloxacin, Neomycin, and Tetracycline Are Neutralizedᰔ Murugan Subbiah,1 Shannon M. Mitchell,2 Jeffrey L. Ullman,4 and Douglas R. Call1,3* Departments of Veterinary Microbiology and Pathology1 and Biological Systems Engineering2 and Paul G. Allen School for Global Animal Health,3 Washington State University, Pullman, Washington 99164, and Department of Agricultural and Biological Engineering, University of Florida, Gainesville, Florida 326114 Received 3 May 2011/Accepted 13 August 2011 It is generally assumed that antibiotic residues in soils select for antibiotic-resistant bacteria. This assump- tion was tested by separately adding 10 different antibiotics (>200 ppm) to three soil-water slurries (silt-loam, sand-loam, and sand; 20% soil [wt/vol]) and incubating mixtures for 24 h at room temperature. The antibiotic activity of the resultant supernatant was assessed by culturing a sensitive Escherichia coli strain in the filter-sterilized supernatant augmented with Luria-Bertani broth. We found striking differences in the abilities of supernatants to suppress growth of the indicator E. coli. Ampicillin, cephalothin, cefoxitin, ceftiofur, and florfenicol supernatants completely inhibited growth while bacterial growth was uninhibited in the presence of neomycin, tetracycline, and ciprofloxacin supernatants. High-performance liquid chromatography (HPLC) analysis demonstrated that cefoxitin and florfenicol were almost completely retained in the supernatants, whereas tetracycline and ciprofloxacin were mostly removed. Antibiotic dissipation in soil, presumably dom- inated by adsorption mechanisms, was sufficient to neutralize 200 ppm of tetracycline; this concentration is considerably higher than reported contamination levels. -

Assessment Report Cyproterone Acetate/Ethinylestradiol (2 Mg/0.035 Mg) Containing Medicinal Products
24 May 2013 EMA/339116/2013 Assessment report cyproterone acetate/ethinylestradiol (2 mg/0.035 mg) containing medicinal products Procedure under Article 107i of Directive 2001/83/EC Procedure number: EMEA/H/A-107i/1357 Assessment Report as adopted by the PRAC with all information of a confidential nature deleted. 7 Westferry Circus ● Canary Wharf ● London E14 4HB ● United Kingdom Telephone +44 (0)20 7418 8400 Facsimile +44 (0)20 7418 8416 E -mail [email protected] Website www.ema.europa.eu An agency of the European Union © European Medicines Agency, 2013. Reproduction is authorised provided the source is acknowledged. Table of contents 1. Background information on the procedure .............................................. 3 2. Scientific discussion ................................................................................ 3 2.1. Clinical aspects .................................................................................................... 4 2.1.1. Clinical safety .................................................................................................... 4 2.1.2. Clinical efficacy ............................................................................................... 22 2.2. Risk minimisation activities .................................................................................. 29 2.3. Product information ............................................................................................ 31 2.4. Benefit-risk assessment ..................................................................................... -

Guidelines for Treatment of Malaria in the United States 1 (Based on Drugs Currently Available for Use in the United States — October 1, 2019)
Guidelines for Treatment of Malaria in the United States 1 (Based on drugs currently available for use in the United States — October 1, 2019) CDC Malaria Hotline: (770) 488-7788 or (855) 856-4713 (toll free) Monday–Friday, 9 am to 5 pm EST; (770) 488-7100 after hours, weekends, and holidays Clinical Diagnosis/ Drug Susceptibility (Based on Recommended Regimen and Adult Dose1 Recommended Regimen and Pediatric Dose1 Plasmodium Species Region Infection Was Acquired) Pediatric dose should NEVER exceed adult dose Uncomplicated malaria/ Chloroquine resistance or unknown A. Artemether-lumefantrine (Coartem™)3,4 P. falciparum, or resistance2 Tablet=20mg artemether/ 120 mg lumefantrine species not identified All malarious regions except those specified A 3-day treatment schedule with a total of 6 oral doses is recommended for both adult and pediatric patients based on as chloroquine sensitive listed in the box weight. The patient should receive the initial dose, followed by the second dose 8 hours later, then 1 dose bid for the If “species not identified” below following 2 days. Dosing as follows: is later diagnosed as P. 5–<15 kg: 1 tablet per dose vivax or P. ovale, please 15–<25 kg: 2 tablets per dose see P. vivax and P. ovale 25–<35 kg: 3 tablets per dose (below) re: treatment with ≥35 kg: 4 tablets per dose primaquine or tafenoquine B. Atovaquone-proguanil (Malarone™)4,5 B. Atovaquone-proguanil (Malarone™)4,5 Adult tablet= 250 mg atovaquone/ 100 mg proguanil Adult tab=250 mg atovaquone/ 100 mg proguanil 4 adult tabs po qd x 3 days Peds tab=62.5 mg atovaquone/ 25 mg proguanil 5–<8 kg: 2 peds tabs po qd x 3 days 8–<10 kg: 3 peds tabs po qd x 3 days 10–<20 kg: 1 adult tab po qd x 3 days 20–<30 kg: 2 adult tabs po qd x 3 days 30–<40 kg: 3 adult tabs po qd x 3 days ≥40 kg: 4 adult tabs po qd x 3 days C.