Phylogenetic Relationships of Kaempferia Plants Based on Inter-Simple Sequence Repeat Fingerprints
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Herbal Products and Their Active Constituents Used Alone and in Combination with Antifungal Drugs Against Drug-Resistant Candida Sp
antibiotics Review Herbal Products and Their Active Constituents Used Alone and in Combination with Antifungal Drugs against Drug-Resistant Candida sp. Anna Herman 1,* and Andrzej Przemysław Herman 2 1 Faculty of Health Sciences, Warsaw School of Engineering and Health, Bitwy Warszawskiej 1920 18 Street, 02-366 Warsaw, Poland 2 Department of Genetic Engineering, The Kielanowski Institute of Animal Physiology and Nutrition, Polish Academy of Sciences, Instytucka 3 Street, 05-110 Jabłonna, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-22-856-70-44; Fax: +48-22-646-34-18 Abstract: Clinical isolates of Candida yeast are the most common cause of opportunistic fungal infections resistant to certain antifungal drugs. Therefore, it is necessary to detect more effective anti- fungal agents that would be successful in overcoming such infections. Among them are some herbal products and their active constituents.The purpose of this review is to summarize the current state of knowledge onherbal products and their active constituents havingantifungal activity against drug- resistant Candida sp. used alone and in combination with antifungal drugs.The possible mechanisms of their action on drug-resistant Candida sp. including (1) inhibition of budding yeast transformation into hyphae; (2) inhibition of biofilm formation; (3) inhibition of cell wall or cytoplasmic membrane biosynthesis; (4) ROS production; and (5) over-expression of membrane transporters will be also described. Citation: Herman, A.; Herman, A.P. Herbal Products and Their Active Keywords: herbalproducts; herbal active constituents; drug-resistant Candida sp.; antifungal drug Constituents Used Alone and in Combination with Antifungal Drugs against Drug-Resistant Candida sp. -

Introduction Common Gynecologic Ailments Could Be Menstrual Period
สมนุ ไพรสำ หรบั โรคสตรที ใี่ ชโ้ ดยหมอพนื้ บำ้ นในจงั หวดั นครนำยก The Use of Medicinal Plants for Gynecologic Ailments by Thai Traditional Folk Healers in Nakhonnayok Province นิพนธ์ต้นฉบ ับ Original Article วรพรรณ สทิ ธถิ าวร1*, ลลิตา วีระเสถียร1 และ ชไมพร อ ้นสว่าง2 Worapan Sitthithaworn1*, Lalita Weerasathien1 and Chamaiporn Onsawang2 1 สาขาเภสชั เวท คณะเภสชั ศาสตร ์ มหาวิทยาลัยศรีนครินทรวิโรฒ อ.องครักษ์ จ.นครนายก 26120 1 Department of Pharmacognosy, Faculty of Pharmacy, Srinakharinwirot University, Ongkarak, 2 งานแพทย์แผนไทย กลุ่มงานคุม้ ครองผูบ้ รโิ ภคและเภสชั สาธารณสขุ ส านักงานสาธารณสขุ จังหวัดนครนายก Nakonnayok 26120, Thailand 2 อ.เมือง จ.นครนายก 26000 Thai Traditional Medicine Unit, Division of Consumer Protection and Public Health Pharmacy, Nakhonnayok Public Health Administration Office, Muang, Nakonnayok 26000, Thailand * Corresponding author: [email protected] * Corresponding author: [email protected] วำรสำรไทยเภสชั ศำสตรแ์ ละวทิ ยำกำรสุขภำพ 2562;14(3):111-121. Thai Pharmaceutical and Health Science Journal 2019;14(3):111-121. บทค ัดย่อ Abstract วัตถุประสงค์: เพื่อระบุสมุนไพรที่หมอพื้นบ้านในจังหวัดนครนายกใช้รักษาโรค Objective: To determine medicinal plants used by folk healers in สตรีในกลุ่มอาการไข้ทับระดู ปวดประจาเดือน ประจาเดือนมาไม่ปกติ และตกขาว Nakhonnayok province for gynecological ailments including pelvic และศึกษาความสัมพันธ์ของสรรพคุณสมุนไพรกับผลการศึกษาฤทธทิ์ างเภสชั inflammatory disease (menstrual fever), dysmenorrhea, oligomenorrhea and วิทยาที่มีรายงานไว้ วิธีการศึกษา: การวิจัยเชิงคุณภาพนี้เก็บข้อมูลโดยการ -

Therapeutic Effects of Bossenbergia Rotunda
International Journal of Science and Research (IJSR) ISSN (Online): 2319-7064 Index Copernicus Value (2015): 78.96 | Impact Factor (2015): 6.391 Therapeutic Effects of Bossenbergia rotunda S. Aishwarya Bachelor of Dental Surgery, Saveetha Dental College and Hospitals Abstract: Boesenbergia rotunda (L.) (Fingerroot), formerly known as Boesenbergia or Kaempferiapandurata (Roxb). Schltr. (Zingiberaceae), is distributed in south-east Asian countries, such as Indonesia, Malaysia and Thailand. The rhizomes of this plant have been used for the treatment of peptic ulcer, as well as colic, oral diseases, urinary disorders, dysentery and inflammation. As people have started to focus more on natural plants species for their curative properties. B. rotunda is a native ingredient in many Asian countries and is used as a condiment in food. It is also used as traditional medicine to treat several illnesses, consumed as traditional tonic especially after childbirth, beauty aid for teenage girls, and as a leukorrhea preventive remedy for women. Its fresh rhizomes are also used to treat inflammatory diseases, in addition to being used as an antifungal, antiparasitic, and aphrodisiac among Thai folks. Moreover, AIDS patients self-medicate themselves with B. rotunda to cure the infection. With the advancement in technology, the ethnomedicinal usages of herbal plants can be explained through in vitro and in vivo studies to prove the activities of the plant extracts. The current state of research on B. rotunda clearly shows that the isolated bioactive compounds have high potential in treating many diseases. Keywords: Zingerberaceae, anti fungal, anti parasitic, Chalcones, flavonoids. 1. Introduction panduratin derivative are prenylated flavonoids from B. pandurata that showed broad range of biological activities, Boesenbergia rotunda is a ginger species that grows in such as strong antibacterial acitivity9-11, anti- inflammatory Southeast Asia, India, Sri Lanka, and Southern China. -

Micropropagation-An in Vitro Technique for the Conservation of Alpinia Galanga
Available online a t www.pelagiaresearchlibrary.com Pelagia Research Library Advances in Applied Science Research, 2014, 5(3):259-263 ISSN: 0976-8610 CODEN (USA): AASRFC Micropropagation-an in vitro technique for the conservation of Alpinia galanga Nongmaithem M. Singh 1, Lukram A. Chanu 1, Yendrembam P. Devi 1, Wahengbam R.C. Singh 2 and Heigrujam B. Singh 2 1DBT-Institutional Biotech Hub, Pettigrew College, Ukhrul, Manipur 2DBT- Institutional Biotech Hub, Deptt. of Biotechnology, S.K. Women’s College, Nambol, Manipur _____________________________________________________________________________________________ ABSTRACT This study was conducted to develop an efficient protocol for mass propagation of Alpinia galanga L. Explants from rhizome buds were cultured on Murashige and Skoog (MS) medium supplemented with 6-Benzylaminopurine (BAP) alone (0 to 5 mg/l) or a combination of BAP (0 to 5 mg/l) and indole 3-acetic acid (IAA) (0 to 2 mg/l). MS medium supplemented with a combination of 5.0 mg/l BAP and 2.0 mg/l IAA or 3.0 mg/l BAP and 0.5 mg/l IAA produced the highest mean number of shoots per explant as compared to other concentrations. The best shoot length was obtained on the medium containing 1.0 mg/l of BAP and 2.0 mg/l IAA. Thus, combined effects of BAP and IAA improved significantly the shoot growth and proliferation. MS medium supplemented with a combination of 5.0 mg/l BAP and 2 mg/l IAA gave the highest number of roots. However, longest roots per explant were obtained with 1.0 mg/l BAP alone. -

C-23 Phytochemical of Kaempferia Plant And
Proceeding of International Conference On Research, Implementation And Education Of Mathematics And Sciences 2014, Yogyakarta State University, 18-20 May 2014 C-23 PHYTOCHEMICAL OF KAEMPFERIA PLANT AND BIOPROSPECTING FOR CANCER TREATMENT Sri Atun Chemistry education Faculty of Mathematical and Natural Science, Yogyakarta State University, Jl. Colombo No. 1 Yogyakarta, Indonesia, 55281 e-mail : [email protected] ABSTRACT Kaempferia genus is perennial member of the Zingiberaceae family and is cultivated in Indonesia and other parts of Southeast Asia. Number of studies has been conducted, providing information related to Kaempferia as antioxidant; antimutagenic; and chemopreventive agent. This paper reports some isolated compounds from this plant, biological activity, and bioprospecting for cancer treatment. Keyword: Cancer treatment; Kaempferia; Zingiberaceae INTRODUCTION Kaempferia is a genus, belong to family of Zingiberaceae. This plant grows in Southeast Asia, India, Sri Lanka, Indonesia, and Southem China. Kaempferia genus sinonim with Boesenbergia genus by Baker. This plant has 8 different botanical names which are Boesenbergia cochinchinensis (Gagnep.) Loes., Boesenbergia pandurata (Roxb.) Schltr., Curcuma rotunda L., Gastrochilus panduratus (Roxb.) Ridl., Gastrochilus rotundus (L.) Alston, Kaempferia cochinchinensis Gagnep., Kaempferia ovate Roscoe, Kaempferia galanga, Kaempferia rotunda, and Kaempferia pandurata Roxb nonetheless it is currently known as Boesenbergia rotunda (L.)Mansf (Tan Eng-Chong, et. al, 2012). The plants grown naturally in damp, shaded parts of the lowland or on hill slopes, as scattered plants or thickets. Economically important species among the plant families, the Zingiberaceae, which are perennial rhizomatous herbs, contain volatile oil and other important compounds of enormous medicinal values (Singh C.B., 2013). Phytochemical and biologycal activities of some species of Kaempferia Phytochemical and biologycal some species of plants of the genus Kaempferia reported by many researchers, among others: 1. -

Vasorelaxant Effect of Boesenbergia Rotunda and Its Active Ingredients
plants Article Vasorelaxant Effect of Boesenbergia rotunda and Its Active Ingredients on an Isolated Coronary Artery 1, 1, 1 1 2 Deepak Adhikari y, Dal-Seong Gong y, Se Hee Oh , Eun Hee Sung , Seung On Lee , Dong-Wook Kim 2, Min-Ho Oak 1,* and Hyun Jung Kim 1,* 1 College of Pharmacy and Natural Medicine Research Institute, Mokpo National University, Muan-gun 58554, Korea; [email protected] (D.A.); [email protected] (D.-S.G.); [email protected] (S.H.O.); [email protected] (E.H.S.) 2 Department of Oriental Medicine Resources, Mokpo National University, Muan-gun 58554, Korea; [email protected] (S.O.L.); [email protected] (D.-W.K.) * Correspondence: [email protected] (M.-H.O.); [email protected] (H.J.K.); Tel.: +82-61-450-2681 (M.-H.O.); +82-61-450-2686 (H.J.K.) The authors contributed to this work equally. y Received: 4 November 2020; Accepted: 27 November 2020; Published: 1 December 2020 Abstract: Cardiovascular diseases are a major cause of death in developed countries. The regulation of vascular tone is a major approach to prevent and ameliorate vascular diseases. As part of our ongoing screening for cardioprotective natural compounds, we investigated the vasorelaxant effect of rhizomes from Boesenbergia rotunda (L.) Mansf. [Boesenbergia pandurata (Roxb.) Schltr.] used as a spice and herbal medicine in Asian countries. The methanol extract of B. rotunda rhizomes (BRE) exhibited significant vasorelaxation effects ex vivo at EC values of 13.4 6.1 µg/mL and 40.9 7.9 µg/mL, 50 ± ± respectively, with and without endothelium in the porcine coronary artery ring. -
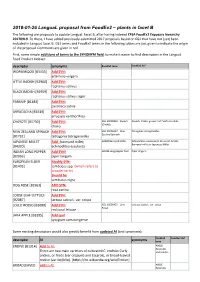
2018-01-26 Langual Proposal from Foodex2 – Plants in Facet B
2018-01-26 LanguaL proposal from FoodEx2 – plants in facet B The following are proposals to update LanguaL Facet B, after having indexed EFSA FoodEx2 Exposure hierarchy 20170919. To these, I have added previously-submitted 2017 proposals based on GS1 that have not (yet) been included in LanguaL facet B. GS1 terms and FoodEx2 terms in the following tables are just given to indicate the origin of the proposal. Comments are given in red. First, some simple additions of terms to the SYNONYM field, to make it easier to find descriptors in the LanguaL Food Product Indexer: descriptor synonyms FoodEx2 term FoodEx2 def WORMWOOD [B3433] Add SYN: artemisia vulgaris LITTLE RADISH [B2960] Add SYN: raphanus sativus BLACK RADISH [B2959] Add SYN: raphanus sativus niger PARSNIP [B1483] Add SYN: pastinaca sativa ARRACACHA [B3439] Add SYN: arracacia xanthorrhiza CHAYOTE [B1730] Add SYN: GS1 10006356 - Squash Squash, Choko, grown from Sechium edule (Choko) choko NEW ZEALAND SPINACH Add SYN: GS1 10006427 - New- Tetragonia tetragonoides Zealand Spinach [B1732] tetragonia tetragonoides JAPANESE MILLET Add : barnyard millet; A000Z Barnyard millet Echinochloa esculenta (A. Braun) H. Scholz, Barnyard millet or Japanese Millet. [B4320] echinochloa esculenta INDIAN LONG PEPPER Add SYN! A019B Long pepper fruit Piper longum [B2956] piper longum EUROPEAN ELDER Modify SYN: [B1403] sambucus spp. (which refers to broader term) Should be sambucus nigra DOG ROSE [B2961] ADD SYN: rosa canina LOOSE LEAF LETTUCE Add SYN: [B2087] lactusa sativa L. var. crispa LOLLO ROSSO [B2088] Add SYN: GS1 10006425 - Lollo Lactuca sativa L. var. crispa Rosso red coral lettuce JAVA APPLE [B3395] Add syn! syzygium samarangense Some existing descriptors would also greatly benefit from updated AI (and synonyms): FoodEx2 FoodEx2 def descriptor AI synonyms term ENDIVE [B1314] Add to AI: A00LD Escaroles There are two main varieties of cultivated C. -

Thai Zingiberaceae : Species Diversity and Their Uses
URL: http://www.iupac.org/symposia/proceedings/phuket97/sirirugsa.html © 1999 IUPAC Thai Zingiberaceae : Species Diversity And Their Uses Puangpen Sirirugsa Department of Biology, Faculty of Science, Prince of Songkla University, Hat Yai, Thailand Abstract: Zingiberaceae is one of the largest families of the plant kingdom. It is important natural resources that provide many useful products for food, spices, medicines, dyes, perfume and aesthetics to man. Zingiber officinale, for example, has been used for many years as spices and in traditional forms of medicine to treat a variety of diseases. Recently, scientific study has sought to reveal the bioactive compounds of the rhizome. It has been found to be effective in the treatment of thrombosis, sea sickness, migraine and rheumatism. GENERAL CHARACTERISTICS OF THE FAMILY ZINGIBERACEAE Perennial rhizomatous herbs. Leaves simple, distichous. Inflorescence terminal on the leafy shoot or on the lateral shoot. Flower delicate, ephemeral and highly modified. All parts of the plant aromatic. Fruit a capsule. HABITATS Species of the Zingiberaceae are the ground plants of the tropical forests. They mostly grow in damp and humid shady places. They are also found infrequently in secondary forest. Some species can fully expose to the sun, and grow on high elevation. DISTRIBUTION Zingiberaceae are distributed mostly in tropical and subtropical areas. The center of distribution is in SE Asia. The greatest concentration of genera and species is in the Malesian region (Indonesia, Malaysia, Singapore, Brunei, the Philippines and Papua New Guinea) *Invited lecture presented at the International Conference on Biodiversity and Bioresources: Conservation and Utilization, 23–27 November 1997, Phuket, Thailand. -

Complete Chloroplast Genomes of Three Medicinal Alpinia Species: Genome Organization, Comparative Analyses and Phylogenetic Relationships in Family Zingiberaceae
plants Article Complete Chloroplast Genomes of Three Medicinal Alpinia Species: Genome Organization, Comparative Analyses and Phylogenetic Relationships in Family Zingiberaceae Dong-Mei Li *, Gen-Fa Zhu *, Ye-Chun Xu, Yuan-Jun Ye and Jin-Mei Liu Guangdong Key Lab of Ornamental Plant Germplasm Innovation and Utilization, Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China; [email protected] (Y.-C.X.); [email protected] (Y.-J.Y.); [email protected] (J.-M.L.) * Correspondence: [email protected] (D.-M.L.); [email protected] (G.-F.Z.); Tel.: +86-20-875-93429 (D.-M.L.) Received: 6 February 2020; Accepted: 20 February 2020; Published: 24 February 2020 Abstract: Alpinia katsumadai (A. katsumadai), Alpinia oxyphylla (A. oxyphylla) and Alpinia pumila (A. pumila), which belong to the family Zingiberaceae, exhibit multiple medicinal properties. The chloroplast genome of a non-model plant provides valuable information for species identification and phylogenetic analysis. Here, we sequenced three complete chloroplast genomes of A. katsumadai, A. oxyphylla sampled from Guangdong and A. pumila, and analyzed the published chloroplast genomes of Alpinia zerumbet (A. zerumbet) and A. oxyphylla sampled from Hainan to retrieve useful chloroplast molecular resources for Alpinia. The five Alpinia chloroplast genomes possessed typical quadripartite structures comprising of a large single copy (LSC, 87,248–87,667 bp), a small single copy (SSC, 15,306–18,295 bp) and a pair of inverted repeats (IR, 26,917–29,707 bp). They had similar gene contents, gene orders and GC contents, but were slightly different in the numbers of small sequence repeats (SSRs) and long repeats. -

Mansf. and Myristica Fragrans Houtt. Against Helicobacter Pylori
Antibacterial activity of Boesenbergia rotunda (L.) Mansf. and Myristica fragrans Houtt. against Helicobacter pylori Sutatip Bhamarapravati1, Siriyaporn Juthapruth2, Warocha Mahachai3 and Gail Mahady4 Abstract Bhamarapravati, S., Juthapruth, S., Mahachai, W. and Mahady, G. Antibacterial activity of Boesenbergia rotunda (L.) Mansf. and Myristica fragrans Houtt. against Helicobacter pylori Songklanakarin J. Sci. Technol., 2006, 28(Suppl. 1) : 157-163 Helicobacter pylori, a gram-negative bacterium, is recognized as the primary etiological agent for the development of gastritis, dyspepsia, peptic ulcer as well as gastric and colon cancer. In developing countries the incidence of H. pylori infection ranges from 50-100%. Two Thai plants, namely Boesenbergia rotunda (L.) Mansf. and Myristica fragrans Houtt., have been used to treat dyspepsia and peptic ulcer in Thai Traditional Medicine. Their crude extracts were previously reported to possess anti- H. pylori activity. This investigation proposed to test previously isolated bioactive compounds from B. rotunda and M. fragrans if they possessed anti- H. pylori activity. Primary cultures of H. pylori from local hospital patients in Thailand were used in the investigation. In vitro anti- H. pylori testing had been performed with pinostrobin and red oil from roots of B. rotunda, and dihydroguaiaretic acid from arils of M. fragrans. Clarithromycin (MIC 120 µg/mL) was used as a positive control. All three compounds showed positive clear zone in agar diffusion test at p<0.05 in all 1Department of Graduate Studies, Faculty of Medicine, Thammasat University, Pathum Thani, 12121 Thai- land. 2M.Sc. student in Microbiology, Faculty of Science, 3MD., Department of Internal Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, 10330 Thailand. -

Complete Chloroplast Genome Sequences of Kaempferia Galanga and Kaempferia Elegans: Molecular Structures and Comparative Analysis
molecules Article Complete Chloroplast Genome Sequences of Kaempferia Galanga and Kaempferia Elegans: Molecular Structures and Comparative Analysis Dong-Mei Li *, Chao-Yi Zhao and Xiao-Fei Liu Guangdong Key Lab of Ornamental Plant Germplasm Innovation and Utilization, Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China; [email protected] (C.-Y.Z.); [email protected] (X.-F.L.) * Correspondence: [email protected]; Tel.: +86-20-875-93429 Received: 27 December 2018; Accepted: 25 January 2019; Published: 29 January 2019 Abstract: Kaempferia galanga and Kaempferia elegans, which belong to the genus Kaempferia family Zingiberaceae, are used as valuable herbal medicine and ornamental plants, respectively. The chloroplast genomes have been used for molecular markers, species identification and phylogenetic studies. In this study, the complete chloroplast genome sequences of K. galanga and K. elegans are reported. Results show that the complete chloroplast genome of K. galanga is 163,811 bp long, having a quadripartite structure with large single copy (LSC) of 88,405 bp and a small single copy (SSC) of 15,812 bp separated by inverted repeats (IRs) of 29,797 bp. Similarly, the complete chloroplast genome of K. elegans is 163,555 bp long, having a quadripartite structure in which IRs of 29,773 bp length separates 88,020 bp of LSC and 15,989 bp of SSC. A total of 111 genes in K. galanga and 113 genes in K. elegans comprised 79 protein-coding genes and 4 ribosomal RNA (rRNA) genes, as well as 28 and 30 transfer RNA (tRNA) genes in K. galanga and K. -

Dictionary of Cultivated Plants and Their Regions of Diversity Second Edition Revised Of: A.C
Dictionary of cultivated plants and their regions of diversity Second edition revised of: A.C. Zeven and P.M. Zhukovsky, 1975, Dictionary of cultivated plants and their centres of diversity 'N -'\:K 1~ Li Dictionary of cultivated plants and their regions of diversity Excluding most ornamentals, forest trees and lower plants A.C. Zeven andJ.M.J, de Wet K pudoc Centre for Agricultural Publishing and Documentation Wageningen - 1982 ~T—^/-/- /+<>?- •/ CIP-GEGEVENS Zeven, A.C. Dictionary ofcultivate d plants andthei rregion so f diversity: excluding mostornamentals ,fores t treesan d lowerplant s/ A.C .Zeve n andJ.M.J ,d eWet .- Wageninge n : Pudoc. -11 1 Herz,uitg . van:Dictionar y of cultivatedplant s andthei r centreso fdiversit y /A.C .Zeve n andP.M . Zhukovsky, 1975.- Me t index,lit .opg . ISBN 90-220-0785-5 SISO63 2UD C63 3 Trefw.:plantenteelt . ISBN 90-220-0785-5 ©Centre forAgricultura l Publishing and Documentation, Wageningen,1982 . Nopar t of thisboo k mayb e reproduced andpublishe d in any form,b y print, photoprint,microfil m or any othermean swithou t written permission from thepublisher . Contents Preface 7 History of thewor k 8 Origins of agriculture anddomesticatio n ofplant s Cradles of agriculture and regions of diversity 21 1 Chinese-Japanese Region 32 2 Indochinese-IndonesianRegio n 48 3 Australian Region 65 4 Hindustani Region 70 5 Central AsianRegio n 81 6 NearEaster n Region 87 7 Mediterranean Region 103 8 African Region 121 9 European-Siberian Region 148 10 South American Region 164 11 CentralAmerica n andMexica n Region 185 12 NorthAmerica n Region 199 Specieswithou t an identified region 207 References 209 Indexo fbotanica l names 228 Preface The aimo f thiswor k ist ogiv e thereade r quick reference toth e regionso f diversity ofcultivate d plants.Fo r important crops,region so fdiversit y of related wild species areals opresented .Wil d species areofte nusefu l sources of genes to improve thevalu eo fcrops .