Towards Predicting Drug-Induced Liver Injury (DILI): Parallel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Medical Review(S) Clinical Review
CENTER FOR DRUG EVALUATION AND RESEARCH APPLICATION NUMBER: 200327 MEDICAL REVIEW(S) CLINICAL REVIEW Application Type NDA Application Number(s) 200327 Priority or Standard Standard Submit Date(s) December 29, 2009 Received Date(s) December 30, 2009 PDUFA Goal Date October 30, 2010 Division / Office Division of Anti-Infective and Ophthalmology Products Office of Antimicrobial Products Reviewer Name(s) Ariel Ramirez Porcalla, MD, MPH Neil Rellosa, MD Review Completion October 29, 2010 Date Established Name Ceftaroline fosamil for injection (Proposed) Trade Name Teflaro Therapeutic Class Cephalosporin; ß-lactams Applicant Cerexa, Inc. Forest Laboratories, Inc. Formulation(s) 400 mg/vial and 600 mg/vial Intravenous Dosing Regimen 600 mg every 12 hours by IV infusion Indication(s) Acute Bacterial Skin and Skin Structure Infection (ABSSSI); Community-acquired Bacterial Pneumonia (CABP) Intended Population(s) Adults ≥ 18 years of age Template Version: March 6, 2009 Reference ID: 2857265 Clinical Review Ariel Ramirez Porcalla, MD, MPH Neil Rellosa, MD NDA 200327: Teflaro (ceftaroline fosamil) Table of Contents 1 RECOMMENDATIONS/RISK BENEFIT ASSESSMENT ......................................... 9 1.1 Recommendation on Regulatory Action ........................................................... 10 1.2 Risk Benefit Assessment.................................................................................. 10 1.3 Recommendations for Postmarketing Risk Evaluation and Mitigation Strategies ........................................................................................................................ -

Research Journal of Pharmaceutical, Biological and Chemical Sciences
ISSN: 0975-8585 Research Journal of Pharmaceutical, Biological and Chemical Sciences Formulation and Evaluation of Floating Drotaverine Hydrochloride Tablets Using Factorial Design Om Prakash*, S Saraf, M Rahman, Neeraj Agnihotri, and Vinay Pathak Department of Industrial chemistry, Integral University, Lucknow. Uttar Pradesh, India. ABSTRACT The main aim of this study was to optimize and evaluate the floating tablets of Drotaverine HCl that prolong the gastric residence time, increasing drug bioavailability and control pain for longer duration by oral administration. A floating drug delivery system(FDDS) was developed using gas forming agent like sodium bicarbonate, citric acid polymers like hydroxypropyl methyl cellulose(HPMC), Sod CMC, Carbopol-934P, PVP K-30. In 32 factorial design amount of HPMC(X1) and gas generating agents(X2) were selected as independent variable and % drug release for 30min,1h, 2h,4h,6h, 8h,12h, 16h , 24h and floating lag time (FLT) were taken as dependent variable. The floating tablet formulations were evaluated for Bulk density (gm/cm3), Tapped density(gm/cm3), Hausner ratio(HR), Carr index, Angle of repose, flow property, assay, in-vitro drug release, hardness, friability,weight variation. The results of in vitro release studies showed that the optimized formulation (F9) could sustain drug release (98.74%) for 24h and remain buoyant for more than 24h. The combination of hydrophilic (HPMC) and hydrophobic (carbopol-934P) polymer provides a better option for 24h release action, bioavailability, stability of tablets at 400C/75%RH, of optimized formulation was carried for one month and no significant change was observed. Keywords: floating drug delivery system(FDDS), Drotaverine HCl, gas generating agents, floating lag time (FLT), hydrophilic (HPMC) and hydrophobic (carbopol-934P) polymer. -
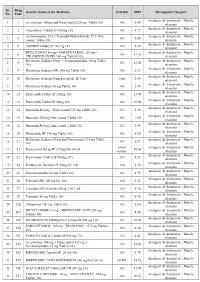
PMBJP Product.Pdf
Sr. Drug Generic Name of the Medicine Unit Size MRP Therapeutic Category No. Code Analgesic & Antipyretic / Muscle 1 1 Aceclofenac 100mg and Paracetamol 325 mg Tablet 10's 10's 8.00 relaxants Analgesic & Antipyretic / Muscle 2 2 Aceclofenac Tablets IP 100mg 10's 10's 4.37 relaxants Acetaminophen 325 + Tramadol Hydrochloride 37.5 film Analgesic & Antipyretic / Muscle 3 4 10's 8.00 coated Tablet 10's relaxants Analgesic & Antipyretic / Muscle 4 5 ASPIRIN Tablets IP 150 mg 14's 14's 2.70 relaxants DICLOFENAC 50 mg+ PARACETAMOL 325 mg+ Analgesic & Antipyretic / Muscle 5 6 10's 11.30 CHLORZOXAZONE 500 mg Tablets 10's relaxants Diclofenac Sodium 50mg + Serratiopeptidase 10mg Tablet Analgesic & Antipyretic / Muscle 6 8 10's 12.00 10's relaxants Analgesic & Antipyretic / Muscle 7 9 Diclofenac Sodium (SR) 100 mg Tablet 10's 10's 6.12 relaxants Analgesic & Antipyretic / Muscle 8 10 Diclofenac Sodium 25mg per ml Inj. IP 3 ml 3 ml 2.00 relaxants Analgesic & Antipyretic / Muscle 9 11 Diclofenac Sodium 50 mg Tablet 10's 10's 2.90 relaxants Analgesic & Antipyretic / Muscle 10 12 Etoricoxilb Tablets IP 120mg 10's 10's 33.00 relaxants Analgesic & Antipyretic / Muscle 11 13 Etoricoxilb Tablets IP 90mg 10's 10's 25.00 relaxants Analgesic & Antipyretic / Muscle 12 14 Ibuprofen 400 mg + Paracetamol 325 mg Tablet 10's 15's 5.50 relaxants Analgesic & Antipyretic / Muscle 13 15 Ibuprofen 200 mg film coated Tablet 10's 10's 1.80 relaxants Analgesic & Antipyretic / Muscle 14 16 Ibuprofen 400 mg film coated Tablet 10's 15's 3.50 relaxants Analgesic & Antipyretic -

)&F1y3x PHARMACEUTICAL APPENDIX to THE
)&f1y3X PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE )&f1y3X PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 3 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. Product CAS No. Product CAS No. ABAMECTIN 65195-55-3 ACTODIGIN 36983-69-4 ABANOQUIL 90402-40-7 ADAFENOXATE 82168-26-1 ABCIXIMAB 143653-53-6 ADAMEXINE 54785-02-3 ABECARNIL 111841-85-1 ADAPALENE 106685-40-9 ABITESARTAN 137882-98-5 ADAPROLOL 101479-70-3 ABLUKAST 96566-25-5 ADATANSERIN 127266-56-2 ABUNIDAZOLE 91017-58-2 ADEFOVIR 106941-25-7 ACADESINE 2627-69-2 ADELMIDROL 1675-66-7 ACAMPROSATE 77337-76-9 ADEMETIONINE 17176-17-9 ACAPRAZINE 55485-20-6 ADENOSINE PHOSPHATE 61-19-8 ACARBOSE 56180-94-0 ADIBENDAN 100510-33-6 ACEBROCHOL 514-50-1 ADICILLIN 525-94-0 ACEBURIC ACID 26976-72-7 ADIMOLOL 78459-19-5 ACEBUTOLOL 37517-30-9 ADINAZOLAM 37115-32-5 ACECAINIDE 32795-44-1 ADIPHENINE 64-95-9 ACECARBROMAL 77-66-7 ADIPIODONE 606-17-7 ACECLIDINE 827-61-2 ADITEREN 56066-19-4 ACECLOFENAC 89796-99-6 ADITOPRIM 56066-63-8 ACEDAPSONE 77-46-3 ADOSOPINE 88124-26-9 ACEDIASULFONE SODIUM 127-60-6 ADOZELESIN 110314-48-2 ACEDOBEN 556-08-1 ADRAFINIL 63547-13-7 ACEFLURANOL 80595-73-9 ADRENALONE -

Acrolein As a Novel Therapeutic Target for Motor and Sensory Deficits in Spinal Cord Injury
[Downloaded free from http://www.nrronline.org on Wednesday, September 11, 2019, IP: 128.210.106.129] NEURAL REGENERATION RESEARCH April 2014,Volume 9,Issue 7 www.nrronline.org SPECIAL ISSUE Acrolein as a novel therapeutic target for motor and sensory deficits in spinal cord injury Jonghyuck Park1, 2, Breanne Muratori2, Riyi Shi1, 2 1 Department of Basic Medical Sciences, College of Veterinary Medicine, Purdue University, West Lafayette, IN, USA 2 Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, USA Abstract Corresponding author: In the hours to weeks following traumatic spinal cord injuries (SCI), biochemical processes are Riyi Shi, M.D., Ph.D., initiated that further damage the tissue within and surrounding the initial injury site: a process Department of Basic Medical Sciences, termed secondary injury. Acrolein, a highly reactive unsaturated aldehyde, has been shown to play College of Veterinary Medicine, Purdue a major role in the secondary injury by contributing significantly to both motor and sensory defi- University, West Lafayette, IN 47907, cits. In particular, efforts have been made to elucidate the mechanisms of acrolein-mediated dam- USA, [email protected]. age at the cellular level and the resulting paralysis and neuropathic pain. In this review, we will highlight the recent developments in the understanding of the mechanisms of acrolein in motor doi:10.4103/1673-5374.131564 and sensory dysfunction in animal models of SCI. We will also discuss the therapeutic benefits of http://www.nrronline.org/ using acrolein scavengers to attenuate acrolein-mediated neuronal damage following SCI. Accepted: 2014-04-08 Key Words: oxidative stress; spinal cord injury; 3-hydrxypropyl mercapturic acid; acrolein-lysine ad- duct; hydralazine Park J, Muratori B, Shi RY. -

328 United States Tariff Commission July 1970 UNITED STATES TARIFF COMMISSION
UNITED STATES TARIFF COMMISSION Washington IMPORTS OF BENZENOID CHEMICALS AND PRODUCTS 1969 United States General Imports of Intermediates, Dyes, Medicinals, Flavor and Perfume Materials, and Other Finished Benzenoid Products Entered on 1969 Under Schedule 4, Part 1, of The Tariff Schedules of the United States TC Publication 328 United States Tariff Commission July 1970 UNITED STATES TARIFF COMMISSION Glenn W. Sutton Bruce E. Clubb Will E. Leonard, Jr. George M. Moore Kenneth R. Mason, Seoretary Address all communications to United States Tariff Commission Washington, D. C. 20436 CONTENTS (Imports under TSUS, Schedule 4, Parts 1B and 1C) Table No. pue_ 1. Benzenoid intermediates: Summary of U.S. general imports entered under Part 1B, TSUS, by competitive status, 1969 4 2. Benzenoid intermediates: U.S. general imports entered under Part 1B, TSUS, by country of origin, 1969 and 1968 3. Benzenoid intermediates: U.S. general iml - orts entered under Part 1B, TSUS, showing competitive status, 1969 4. Finished benzenoid products: Summary of U.S.general . im- ports entered under Part 1C, TSUS, by competitive status, 1969 24 5. Finished benzenoid products: U.S. general imports entered under Part 1C, TSUS, by country of origin, 1969 and 1968 25 6. Finished benzenoid products: Summary of U.S. general imports entered under Part 1C, TSUS, by major groups and competitive status, 1969 27 7. Benzenoid dyes: U.S. general imports entered under Part 1C, TSUS, by class of application, and competitive status, 1969-- 30 8. Benzenoid dyes: U.S. general imports entered under Part 1C, TSUS, by country of origin, 1969 compared with 1968 31 9. -

(12) United States Patent (10) Patent No.: US 6,784,177 B2 Cohn Et Al
USOO6784177B2 (12) United States Patent (10) Patent No.: US 6,784,177 B2 Cohn et al. (45) Date of Patent: Aug. 31, 2004 (54) METHODS USING HYDRALAZINE Massie et al., The American Journal of Cardiology, COMPOUNDS AND SOSORBIDE 40:794-801 (1977). DINTRATE OR ISOSORBIDE Kaplan et al., Annals of Internal Medicine, 84:639-645 MONONTRATE (1976). Bauer et al., Circulation, 84(1):35-39 (1991). (75) Inventors: Jay N. Cohn, Minneapolis, MN (US); The SOLVD Investigators, The New England Journal of Medicine, 327(10):685–691 (1992). Peter Carson, Chevy Chase, MD (US) Ziesche et al., Circulation, 87(6):VI56-VI64 (1993). Rector et al., Circulation, 87(6):VI71-VI77 (1993). (73) Assignee: Nitro Med, Inc., Bedford, MA (US) Carson et al., Journal of Cardiac Failure, 5(3):178-187 (Sep. 10, 1999). (*) Notice: Subject to any disclaimer, the term of this Dries et al, The New England Journal of Medicine, patent is extended or adjusted under 35 340(8):609-616 (Feb. 25, 1999). U.S.C. 154(b) by 18 days. Freedman et al, Drugs, 54 (Supplement 3):41-50 (1997). Sherman et al., Cardiologia, 42(2):177-187 (1997). (21) Appl. No.: 10/210,113 Biegelson et al., Coronary Artery Disease, 10:241-256 (1999). (22) Filed: Aug. 2, 2002 Rudd et al, Am. J. Physiol., 277(46):H732–H739 (1999). (65) Prior Publication Data Hammerman et al, Am. J. Physiol., 277(46):H1579–1592 (1999). US 2004/0023967 A1 Feb. 5, 2004 LoScalzo et al., Transactions of the American and Climato logical ASS., 111:158-163 (2000). -

Minutes of PRAC Meeting on 09-12 July 2018
6 September 2018 EMA/PRAC/576790/2018 Inspections, Human Medicines Pharmacovigilance and Committees Division Pharmacovigilance Risk Assessment Committee (PRAC) Minutes of the meeting on 09-12 July 2018 Chair: June Raine – Vice-Chair: Almath Spooner Health and safety information In accordance with the Agency’s health and safety policy, delegates were briefed on health, safety and emergency information and procedures prior to the start of the meeting. Disclaimers Some of the information contained in the minutes is considered commercially confidential or sensitive and therefore not disclosed. With regard to intended therapeutic indications or procedure scope listed against products, it must be noted that these may not reflect the full wording proposed by applicants and may also change during the course of the review. Additional details on some of these procedures will be published in the PRAC meeting highlights once the procedures are finalised. Of note, the minutes are a working document primarily designed for PRAC members and the work the Committee undertakes. Note on access to documents Some documents mentioned in the minutes cannot be released at present following a request for access to documents within the framework of Regulation (EC) No 1049/2001 as they are subject to on- going procedures for which a final decision has not yet been adopted. They will become public when adopted or considered public according to the principles stated in the Agency policy on access to documents (EMA/127362/2006, Rev. 1). 30 Churchill Place ● Canary Wharf ● London E14 5EU ● United Kingdom Telephone +44 (0)20 3660 6000 Facsimile +44 (0)20 3660 5555 Send a question via our website www.ema.europa.eu/contact An agency of the European Union © European Medicines Agency, 2018. -
![Ehealth DSI [Ehdsi V2.2.2-OR] Ehealth DSI – Master Value Set](https://docslib.b-cdn.net/cover/8870/ehealth-dsi-ehdsi-v2-2-2-or-ehealth-dsi-master-value-set-1028870.webp)
Ehealth DSI [Ehdsi V2.2.2-OR] Ehealth DSI – Master Value Set
MTC eHealth DSI [eHDSI v2.2.2-OR] eHealth DSI – Master Value Set Catalogue Responsible : eHDSI Solution Provider PublishDate : Wed Nov 08 16:16:10 CET 2017 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 1 of 490 MTC Table of Contents epSOSActiveIngredient 4 epSOSAdministrativeGender 148 epSOSAdverseEventType 149 epSOSAllergenNoDrugs 150 epSOSBloodGroup 155 epSOSBloodPressure 156 epSOSCodeNoMedication 157 epSOSCodeProb 158 epSOSConfidentiality 159 epSOSCountry 160 epSOSDisplayLabel 167 epSOSDocumentCode 170 epSOSDoseForm 171 epSOSHealthcareProfessionalRoles 184 epSOSIllnessesandDisorders 186 epSOSLanguage 448 epSOSMedicalDevices 458 epSOSNullFavor 461 epSOSPackage 462 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 2 of 490 MTC epSOSPersonalRelationship 464 epSOSPregnancyInformation 466 epSOSProcedures 467 epSOSReactionAllergy 470 epSOSResolutionOutcome 472 epSOSRoleClass 473 epSOSRouteofAdministration 474 epSOSSections 477 epSOSSeverity 478 epSOSSocialHistory 479 epSOSStatusCode 480 epSOSSubstitutionCode 481 epSOSTelecomAddress 482 epSOSTimingEvent 483 epSOSUnits 484 epSOSUnknownInformation 487 epSOSVaccine 488 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 3 of 490 MTC epSOSActiveIngredient epSOSActiveIngredient Value Set ID 1.3.6.1.4.1.12559.11.10.1.3.1.42.24 TRANSLATIONS Code System ID Code System Version Concept Code Description (FSN) 2.16.840.1.113883.6.73 2017-01 A ALIMENTARY TRACT AND METABOLISM 2.16.840.1.113883.6.73 2017-01 -

Pharmaceutical Appendix to the Tariff Schedule 2
Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACIDUM LIDADRONICUM 63132-38-7 ABAFUNGIN 129639-79-8 ACIDUM SALCAPROZICUM 183990-46-7 ABAMECTIN 65195-55-3 ACIDUM SALCLOBUZICUM 387825-03-8 ABANOQUIL 90402-40-7 ACIFRAN 72420-38-3 ABAPERIDONUM 183849-43-6 ACIPIMOX 51037-30-0 ABARELIX 183552-38-7 ACITAZANOLAST 114607-46-4 ABATACEPTUM 332348-12-6 ACITEMATE 101197-99-3 ABCIXIMAB 143653-53-6 ACITRETIN 55079-83-9 ABECARNIL 111841-85-1 ACIVICIN 42228-92-2 ABETIMUSUM 167362-48-3 ACLANTATE 39633-62-0 ABIRATERONE 154229-19-3 ACLARUBICIN 57576-44-0 ABITESARTAN 137882-98-5 ACLATONIUM NAPADISILATE 55077-30-0 ABLUKAST 96566-25-5 ACODAZOLE 79152-85-5 ABRINEURINUM 178535-93-8 ACOLBIFENUM 182167-02-8 ABUNIDAZOLE 91017-58-2 ACONIAZIDE 13410-86-1 ACADESINE 2627-69-2 ACOTIAMIDUM 185106-16-5 ACAMPROSATE 77337-76-9 -

Systematic Evidence Review from the Blood Pressure Expert Panel, 2013
Managing Blood Pressure in Adults Systematic Evidence Review From the Blood Pressure Expert Panel, 2013 Contents Foreword ............................................................................................................................................ vi Blood Pressure Expert Panel ..............................................................................................................vii Section 1: Background and Description of the NHLBI Cardiovascular Risk Reduction Project ............ 1 A. Background .............................................................................................................................. 1 Section 2: Process and Methods Overview ......................................................................................... 3 A. Evidence-Based Approach ....................................................................................................... 3 i. Overview of the Evidence-Based Methodology ................................................................. 3 ii. System for Grading the Body of Evidence ......................................................................... 4 iii. Peer-Review Process ....................................................................................................... 5 B. Critical Question–Based Approach ........................................................................................... 5 i. How the Questions Were Selected ................................................................................... 5 ii. Rationale for the Questions -

Diagnosis and Treatment of Gouty Arthritis JOHN H
Diagnosis and Treatment of Gouty Arthritis JOHN H. TALBOTT, M.D., Buffalo. New York AN ADEQUATE UNDERSTANDING of the approach to the * The characteristic phenomena of acute gouty diagnosis of a clinical entity and an effective regi- arthritis are'acute arthritis in a middle-aged men of therapy should be based upon accepted con- male, associated with serum uric acid above 6 cepts of normal and abnormal function. Diseases mg. per 100 cc. and a satisfactory response to that are identified as metabolic disturbances serve as coichicine. Roentgenographically observable good examples. Although gout is not the example par changes do not occur early. excellence, a clinical discussion of this dyscrasia In recent years uric acid metabolism has may be developed from accepted physiological and been studied by means of isotope techniques biochemical phenomena. utilizing labeled substances. Uric acid is ex- The clinical finding of acute arthritis associated creted in relatively constant amounts by humans with the cardinal signs of inflammation in a male and is little affected by variations in dietary should suggest gout as one possibility. If it is pres- intake, except for purine or nucleic acid sub- ent the amount of uric acid in the serum will be stances. Persons with gout have a greater total above 6 mg. per 100 cc., and if -a "full course" of amount of uric acid and a lower turnover than colchicine is administered, the acute articular symp- normal persons. toms should subside. These items-a characteristic In the treatment of acute affacks of gout col- clinical picture, an elevation of serum urate and a chicine is still the most practical single drug, satisfactory response to colchicine-constitute the even though its pharmacologic action remains triad of acute gouty arthritis.