Pub . No .: US 2021/0010039 A1 Lim Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research Article Antimicrobial and Antioxidant Properties of a Bacterial
Hindawi International Journal of Microbiology Volume 2020, Article ID 9483670, 11 pages https://doi.org/10.1155/2020/9483670 Research Article Antimicrobial and Antioxidant Properties of a Bacterial Endophyte, Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum Seeds Mampolelo M. Photolo ,1 Vuyo Mavumengwana ,2 Lungile Sitole ,1 and Matsobane G. Tlou 3 1Department of Biochemistry, Faculty of Science, University of Johannesburg, Auckland Park Campus, Johannesburg, South Africa 2DST-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Tygerberg Campus, Cape Town, South Africa 3Department of Biochemistry, School of Physical and Chemical Sciences, Faculty of Natural and Agricultural Sciences, North-West University, Mafikeng Campus, South Africa Correspondence should be addressed to Matsobane G. Tlou; [email protected] Received 17 September 2019; Accepted 21 December 2019; Published 25 February 2020 Academic Editor: Karl Drlica Copyright © 2020 Mampolelo M. Photolo et al. -is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -is study reports on the isolation and identification of Methylobacterium radiotolerans MAMP 4754 from the seeds of the medicinal plant, Combretum -

1 Supplementary Figures 1 2 Fine-Scale Adaptations To
1 SUPPLEMENTARY FIGURES 2 3 FINE-SCALE ADAPTATIONS TO ENVIRONMENTAL VARIATION AND GROWTH 4 STRATEGIES DRIVE PHYLLOSPHERE METHYLOBACTERIUM DIVERSITY. 5 6 Jean-Baptiste Leducq1,2,3, Émilie Seyer-Lamontagne1, Domitille Condrain-Morel2, Geneviève 7 Bourret2, David Sneddon3, James A. Foster3, Christopher J. Marx3, Jack M. Sullivan3, B. Jesse 8 Shapiro1,4 & Steven W. Kembel2 9 10 1 - Université de Montréal 11 2 - Université du Québec à Montréal 12 3 – University of Idaho 13 4 – McGill University 14 1 15 Figure S1 - Validation of Methylobacterium group A subdivisions in nine clades. Distribution 16 of pairwise nucleotide similarity (PS) within and among Methylobacterium clades and 17 Microvirga (outgroup) estimated from the complete rpoB nucleotide sequence (4064 bp). PS was 18 calculated in MEGA7 (1, 2) from aligned nucleotide sequences as PS = 1-pdistance. 19 2 A1 A2 A3 A4 A5 A6 Within clades A9 B C Within Methylobacterium among A1−A9 between A and B between A and C between B and C within Microvirga between Methylobacterium and Microvirga 0.75 0.80 0.85 0.90 0.95 1.00 PS 20 Figure S2 - ASV rarefaction on diversity assessed by 16s barcoding in 46 phyllosphere 21 samples, 1 positive control (METH community) and 1 negative controls. a) Diversity in 22 METH community (positive control) and rare ASVs threshold definition. The METH community 23 consisted in mixed genomic DNAs from 18 Methylobacterium isolates representative of diversity 24 isolated in SBL and MSH in 2017 and 2018 (Table S6; Table S10), one Escherichia coli strain 25 and one Sphingomonas sp. isolate from MSH in 2017 (isolate DNA022; Table S6), also probably 26 contaminated with Thermotoga sp. -

Antimicrobial and Antioxidant Properties of a Bacterial Endophyte, Methylobacterium Radiotolerans MAMP 4754, Isolated from Combretum Erythrophyllum Seeds
Hindawi International Journal of Microbiology Volume 2020, Article ID 9483670, 11 pages https://doi.org/10.1155/2020/9483670 Research Article Antimicrobial and Antioxidant Properties of a Bacterial Endophyte, Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum Seeds Mampolelo M. Photolo ,1 Vuyo Mavumengwana ,2 Lungile Sitole ,1 and Matsobane G. Tlou 3 1Department of Biochemistry, Faculty of Science, University of Johannesburg, Auckland Park Campus, Johannesburg, South Africa 2DST-NRF Centre of Excellence for Biomedical Tuberculosis Research, South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Tygerberg Campus, Cape Town, South Africa 3Department of Biochemistry, School of Physical and Chemical Sciences, Faculty of Natural and Agricultural Sciences, North-West University, Mafikeng Campus, South Africa Correspondence should be addressed to Matsobane G. Tlou; [email protected] Received 17 September 2019; Accepted 21 December 2019; Published 25 February 2020 Academic Editor: Karl Drlica Copyright © 2020 Mampolelo M. Photolo et al. -is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -is study reports on the isolation and identification of Methylobacterium radiotolerans MAMP 4754 from the seeds of the medicinal plant, Combretum -

Downloaded from Genbank
Methylotrophs and Methylotroph Populations for Chloromethane Degradation Françoise Bringel1*, Ludovic Besaury2, Pierre Amato3, Eileen Kröber4, Stefen Kolb4, Frank Keppler5,6, Stéphane Vuilleumier1 and Thierry Nadalig1 1Université de Strasbourg UMR 7156 UNISTR CNRS, Molecular Genetics, Genomics, Microbiology (GMGM), Strasbourg, France. 2Université de Reims Champagne-Ardenne, Chaire AFERE, INR, FARE UMR A614, Reims, France. 3 Université Clermont Auvergne, CNRS, SIGMA Clermont, ICCF, Clermont-Ferrand, France. 4Microbial Biogeochemistry, Research Area Landscape Functioning – Leibniz Centre for Agricultural Landscape Research – ZALF, Müncheberg, Germany. 5Institute of Earth Sciences, Heidelberg University, Heidelberg, Germany. 6Heidelberg Center for the Environment HCE, Heidelberg University, Heidelberg, Germany. *Correspondence: [email protected] htps://doi.org/10.21775/cimb.033.149 Abstract characterized ‘chloromethane utilization’ (cmu) Chloromethane is a halogenated volatile organic pathway, so far. Tis pathway may not be representa- compound, produced in large quantities by terres- tive of chloromethane-utilizing populations in the trial vegetation. Afer its release to the troposphere environment as cmu genes are rare in metagenomes. and transport to the stratosphere, its photolysis con- Recently, combined ‘omics’ biological approaches tributes to the degradation of stratospheric ozone. A with chloromethane carbon and hydrogen stable beter knowledge of chloromethane sources (pro- isotope fractionation measurements in microcosms, duction) and sinks (degradation) is a prerequisite indicated that microorganisms in soils and the phyl- to estimate its atmospheric budget in the context of losphere (plant aerial parts) represent major sinks global warming. Te degradation of chloromethane of chloromethane in contrast to more recently by methylotrophic communities in terrestrial envi- recognized microbe-inhabited environments, such ronments is a major underestimated chloromethane as clouds. -

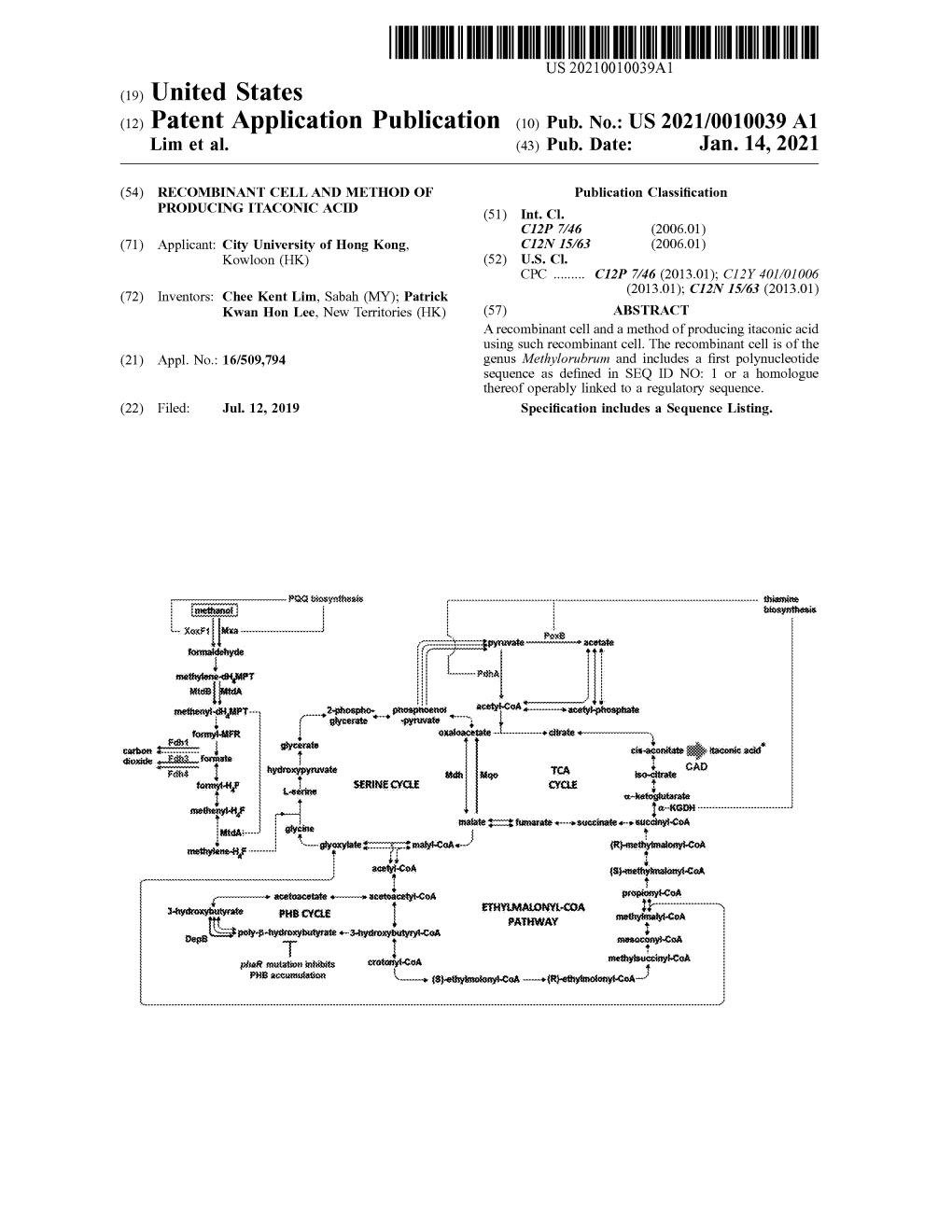
Designing and Engineering Methylorubrum Extorquens AM1 for Itaconic Acid Production
fmicb-10-01027 May 8, 2019 Time: 14:44 # 1 ORIGINAL RESEARCH published: 09 May 2019 doi: 10.3389/fmicb.2019.01027 Designing and Engineering Methylorubrum extorquens AM1 for Itaconic Acid Production Chee Kent Lim1, Juan C. Villada1, Annie Chalifour2†, Maria F. Duran1, Hongyuan Lu1† and Patrick K. H. Lee1* 1 School of Energy and Environment, City University of Hong Kong, Hong Kong, China, 2 Department of Chemistry, City Edited by: University of Hong Kong, Hong Kong, China Sabine Kleinsteuber, Helmholtz Centre for Environmental Research (UFZ), Germany Methylorubrum extorquens (formerly Methylobacterium extorquens) AM1 is a Reviewed by: methylotrophic bacterium with a versatile lifestyle. Various carbon sources including Volker Döring, acetate, succinate and methanol are utilized by M. extorquens AM1 with the latter Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), being a promising inexpensive substrate for use in the biotechnology industry. Itaconic France acid (ITA) is a high-value building block widely used in various industries. Given Norma Cecilia Martinez-Gomez, that no wildtype methylotrophic bacteria are able to utilize methanol to produce Michigan State University, United States ITA, we tested the potential of M. extorquens AM1 as an engineered host for this *Correspondence: purpose. In this study, we successfully engineered M. extorquens AM1 to express Patrick K. H. Lee a heterologous codon-optimized gene encoding cis-aconitic acid decarboxylase. The [email protected] engineered strain produced ITA using acetate, succinate and methanol as the carbon †Present address: Annie Chalifour, feedstock. The highest ITA titer in batch culture with methanol as the carbon source Swiss Federal Institute of Aquatic was 31.6 ± 5.5 mg/L, while the titer and productivity were 5.4 ± 0.2 mg/L and Science and Technology (EAWAG), 0.056 ± 0.002 mg/L/h, respectively, in a scaled-up fed-batch bioreactor under Überlandstrasse, Dübendorf, Switzerland 60% dissolved oxygen saturation. -

(REE)-Dependent Growth of Pseudomonas Putida KT2440
Rare Earth Element (REE)-Dependent Growth of Pseudomonas putida KT2440 Relies on the ABC-Transporter PedA1A2BC and Is Influenced by Iron Availability Matthias Wehrmann, Charlotte Berthelot, Patrick Billard, Janosch Klebensberger To cite this version: Matthias Wehrmann, Charlotte Berthelot, Patrick Billard, Janosch Klebensberger. Rare Earth El- ement (REE)-Dependent Growth of Pseudomonas putida KT2440 Relies on the ABC-Transporter PedA1A2BC and Is Influenced by Iron Availability. Frontiers in Microbiology, Frontiers Media, 2019, 10, 10.3389/fmicb.2019.02494. hal-02389228 HAL Id: hal-02389228 https://hal.archives-ouvertes.fr/hal-02389228 Submitted on 8 Jan 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial| 4.0 International License fmicb-10-02494 November 1, 2019 Time: 15:18 # 1 ORIGINAL RESEARCH published: 31 October 2019 doi: 10.3389/fmicb.2019.02494 Rare Earth Element (REE)-Dependent Growth of Pseudomonas putida KT2440 Relies on the ABC-Transporter PedA1A2BC and Is Influenced by Iron Availability Matthias Wehrmann1, Charlotte Berthelot2,3, Patrick Billard2,3* and Janosch Klebensberger1* 1 Edited by: Department of Technical Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 2 Hari S. -

Taxonomic and Functional Characterization Of
TAXONOMIC AND FUNCTIONAL CHARACTERIZATION OF MICROBIAL COMMUNITIES LINKED TO CHLORINATED SOLVENT, 1,4-DIOXANE AND RDX BIODEGRADATION IN GROUNDWATER AND SOIL MICROCOSMS By Hongyu Dang A DISSERTATION Submitted to Michigan State University in partial fulfillment of the requirements for the degree of Environmental Engineering—Doctor of Philosophy 2021 ABSTRACT TAXONOMIC AND FUNCTIONAL CHARACTERIZATION OF MICROBIAL COMMUNITIES LINKED TO CHLORINATED SOLVENT, 1,4-DIOXANE AND RDX BIODEGRADATION IN GROUNDWATER AND SOIL MICROCOSMS By Hongyu Dang Bioremediation is becoming increasing popular for the remediation of sites contaminated with a range of different contaminants. Molecular methods such as 16S rRNA gene amplicon sequencing, shotgun sequencing, and high throughput quantitative PCR offer much potential for examining the microorganisms and functional genes associated with contaminant biodegradation, which can provide critical additional lines of evidence for effective site remediation. In this work, the first project examined the taxonomic and functional biomarkers associated with chlorinated solvent and 1,4-dioxane biodegradation in groundwater from five contaminated sites. Each site had previously been bioaugmented with the commercially available dechlorinating mixed culture, SDC-9. The results highlighted the occurrence of numerous genera previously linked to chlorinated solvent biodegradation. The functional gene analysis indicated two reductive dehalogenase genes (vcrA and tceA) from Dehalococcoides mccartyi were abundant. Additionally, aerobic and anaerobic biomarkers for the biodegradation of various chlorinated compounds were observed across all sites. The approach used (shotgun sequencing) is advantageous over many other methods because an unlimited number of functional genes can be examined and a more complete picture of the functional abilities of microbial communities can be depicted. Another research project evaluated the functional genes and species associated with RDX biodegradation at a RDX contaminated Navy site where biostimulation had been adopted. -

Lanthanide Transport, Storage, and Beyond: Genes and Processes Contributing to Xoxf Function In
bioRxiv preprint doi: https://doi.org/10.1101/647677; this version posted May 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Lanthanide transport, storage, and beyond: genes and processes contributing to XoxF function in 2 Methylorubrum extorquens AM1 3 4 Paula Roszczenko-Jasińska,a,£ Huong N. Vu,b,*,£ Gabriel A. Subuyuj,b,* Ralph Valentine 5 Crisostomo,b,* James Cai,b Charumathi Raghuraman,b Elena M. Ayala,b Erik J. Clippard,b 6 Nicholas F. Lien,b Richard T. Ngo,b Fauna Yarza,b,* Caitlin A. Hoeber,b Norma C. Martinez- 7 Gomez,a# Elizabeth Skovranb# 8 9 10 aDepartment of Microbiology and Molecular Genetics, Michigan State University, East Lansing, 11 USA 12 bDepartment of Biological Sciences, San José State University, San José, California, USA 13 14 Running Head: Lanthanide transport and storage 15 £ Both authors contributed equally and share first authorship 16 # Address correspondence to: Elizabeth Skovran, [email protected] and N. Cecilia 17 Martinez-Gomez, [email protected] 18 * Present address: HNV: Department of Microbiology, University of Georgia, Athens, Georgia, 19 USA; GAS: Department of Microbiology and Molecular Genetics, University of California at 20 Davis, Davis, California, USA, RVC: Molecular Biology Institute, University of California at 21 Los Angeles, Los Angeles, California, USA; FY: Department of Biochemistry and Biophysics, 22 University of California at San Francisco, San Francisco, California, USA bioRxiv preprint doi: https://doi.org/10.1101/647677; this version posted May 24, 2019. -

Lanthanide-Dependent Methylotrophic Pathway in Methylobacterium Aquaticum Strain 22A
Lanthanide-dependent methylotrophic pathway in Methylobacterium aquaticum strain 22A 2020, September Patcha Yanpirat Graduate School of Environmental and Life Science (Doctor’s Course) OKAYAMA UNIVERSITY Table of Contents Abstract .................................................................................................................................. i Abbreviations ......................................................................................................................... iii 1. Introduction ........................................................................................................................ 1 1.1. Methanol and methylotrophs .............................................................................. 1 1.2. Methylotrophy in Methylobacterium species...................................................... 2 1.3. Role of lanthanides in methylotrophy ................................................................. 3 1.4. Methylobacterium aquaticum strain 22A............................................................ 6 1.5. Formaldehyde oxidation in Ln-dependent methylotrophy in Methylobacterium aquaticum strain 22A ....................................................... 7 2. Materials and Methods ....................................................................................................... 9 2.1. Strain and culture conditions .............................................................................. 9 2.2. Construction of formaldehyde oxidation gene deletion mutants ....................... -

2—Impact of Environmental Changes on the Microbial Community
Dynamics of a methanol-fed marine denitrifying biofilm: 2—impact of environmental changes on the microbial community Richard Villemur1, Geneviève Payette1, Valérie Geoffroy2, Florian Mauffrey3 and Christine Martineau4 1 INRS-Centre Armand-Frappier Santé et Biotechnologie, Laval, Québec, Canada 2 Lallemand, Montréal, Québec, Canada 3 Université de Genève, Geneva, Switzerland 4 Laurentian Forestry Centre, Québec, Canada ABSTRACT Background. The biofilm of a methanol-fed, marine denitrification system is composed of a multi-species microbial community, among which Hyphomicrobium nitrativorans and Methylophaga nitratireducenticrescens are the principal bacteria involved in the denitrifying activities. To assess its resilience to environmental changes, the biofilm was cultivated in artificial seawater (ASW) under anoxic conditions and exposed to a range of specific environmental conditions. We previously reported the impact of these changes on the denitrifying activities and the co-occurrence of H. nitrativorans strain NL23 and M. nitratireducenticrescens in the biofilm cultures. Here, we report the impact of these changes on the dynamics of the overall microbial community of the denitrifying biofilm. Methods. The original biofilm (OB) taken from the denitrification system was cultivated in ASW under anoxic conditions with a range of NaCl concentrations, and with four combinations of nitrate/methanol concentrations and temperatures. The OB was also cultivated in the commercial Instant Ocean seawater (IO). The bacterial Submitted 17 April 2019 diversity of the biofilm cultures and the OB was determined by 16S ribosomal RNA Accepted 12 July 2019 gene sequences. Culture approach was used to isolate other denitrifying bacteria from Published 13 August 2019 the biofilm cultures. The metatranscriptomes of selected biofilm cultures were derived, Corresponding author along with the transcriptomes of planktonic pure cultures of H. -
Methylobacterium Sp. 2A Is a Plant Growth-Promoting Rhizobacteria That Has the Potential to Improve Potato Crop Yield Under Adverse
ORIGINAL RESEARCH published: 14 February 2020 doi: 10.3389/fpls.2020.00071 Methylobacterium sp. 2A Is a Plant Growth-Promoting Rhizobacteria That Has the Potential to Improve Potato Crop Yield Under Adverse Edited by: Conditions Briardo Llorente, 1 1† 2 3 Macquarie University, Cecilia Eugenia María Grossi , Elisa Fantino , Federico Serral , Myriam Sara Zawoznik , Australia Darío Augusto Fernandez Do Porto 2 and Rita María Ulloa 1,4* Reviewed by: 1 Laboratorio de Transducción de Señales en Plantas, Instituto de Investigaciones en Ingeniería Genética y Biología Vasvi Chaudhry, Molecular (INGEBI), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Ciudad Autónoma de Buenos University of Tübingen, Aires, Argentina, 2 Plataforma de Bioinformática Argentina, Instituto de Cálculo, Ciudad Universitaria, Facultad de Ciencias Germany Exactas y Naturales, Universidad de Buenos Aires (UBA), Ciudad Autónoma de Buenos Aires, Argentina, 3 Cátedra de Nurettin Sahin, Química Biológica Vegetal, Departamento de Química Biológica, Facultad de Farmacia y Bioquímica, Universidad de Buenos Mugla Sitki Kocman University, Aires (UBA), Ciudad Autónoma de Buenos Aires, Argentina, 4 Departamento de Química Biológica, Universidad de Buenos Turkey Aires (UBA), Ciudad Autónoma de Buenos Aires, Argentina *Correspondence: Rita María Ulloa [email protected]; A Gram-negative pink-pigmented bacillus (named 2A) was isolated from Solanum [email protected] tuberosum L. cv. Desirée plants that were strikingly more developed, presented † Present address: increased root hair density, and higher biomass than other potato lines of the same Elisa Fantino, Laboratoire de Recherche Sur le age. The 16S ribosomal DNA sequence, used for comparative gene sequence analysis, Métabolisme Spécialisé Végétal, indicated that strain 2A belongs to the genus Methylobacterium. -

Methylobacterium Ajmalii Sp. Nov., Isolated from the International Space Station
fmicb-12-639396 March 12, 2021 Time: 20:27 # 1 ORIGINAL RESEARCH published: 15 March 2021 doi: 10.3389/fmicb.2021.639396 Methylobacterium ajmalii sp. nov., Isolated From the International Space Station Swati Bijlani1, Nitin K. Singh2, V. V. Ramprasad Eedara3, Appa Rao Podile3, Christopher E. Mason4, Clay C. C. Wang1* and Kasthuri Venkateswaran2* 1 Department of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, Los Angeles, CA, United States, 2 Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA, United States, 3 Department of Plant Science, School of Life Sciences, University of Hyderabad, Hyderabad, India, 4 WorldQuant Initiative for Quantitative Prediction, Weill Cornell Medicine, New York, NY, United States Four strains belonging to the family of Methylobacteriaceae were isolated from different locations on the International Space Station (ISS) across two consecutive flights. Of these, three were identified as Gram-negative, rod-shaped, catalase-positive, oxidase- positive, motile bacteria, designated as IF7SW-B2T, IIF1SW-B5, and IIF4SW-B5, whereas the fourth was identified as Methylorubrum rhodesianum. The sequence similarity of these three ISS strains, designated as IF7SW-B2T, IIF1SW-B5, and IIF4SW- Edited by: Brian P. Hedlund, B5, was <99.4% for 16S rRNA genes and <97.3% for gyrB gene, with the closest University of Nevada, Las Vegas, being Methylobacterium indicum SE2.11T. Furthermore, the multi-locus sequence United States analysis placed these three ISS strains in the same clade of M. indicum. The average Reviewed by: Jian-Yu Jiao, nucleotide identity (ANI) values of these three ISS strains were <93% and digital DNA- Sun Yat-sen University, China DNA hybridization (dDDH) values were <46.4% with any described Methylobacterium En-Min Zhou, species.