The Drosophila Dbf4 Ortholog Chiffon Forms a Complex with Gcn5 That Is Necessary for Histone Acetylation and Viability Eliana F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mcm2 Is a Target of Regulation by Cdc7–Dbf4 During the Initiation of DNA Synthesis
Downloaded from genesdev.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis Ming Lei,1 Yasuo Kawasaki,1,2 Michael R. Young,1,3 Makoto Kihara,2 Akio Sugino,2 and Bik K. Tye1,4 1Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, New York 14853-2703 USA; 2Research Institute for Microbial Diseases, Osaka University, Osaka 565, Japan The initiation of DNA synthesis is an important cell cycle event that defines the beginning of S phase. This critical event involves the participation of proteins whose functions are regulated by cyclin dependent protein kinases (Cdks). The Mcm2–7 proteins are a family of six conserved proteins that are essential for the initiation of DNA synthesis in all eukaryotes. In Saccharomyces cerevisiae, members of the Mcm2–7 family undergo cell cycle-specific phosphorylation. Phosphorylation of Mcm proteins at the beginning of S phase coincides with the removal of these proteins from chromatin and the onset of DNA synthesis. In this study, we identified DBF4, which encodes the regulatory subunit of a Cdk-like protein kinase Cdc7–Dbf4, in a screen for second site suppressors of mcm2-1. The dbf4 suppressor mutation restores competence to initiate DNA synthesis to the mcm2-1 mutant. Cdc7–Dbf4 interacts physically with Mcm2 and phosphorylates Mcm2 and three other members of the Mcm2–7 family in vitro. Blocking the kinase activity of Cdc7–Dbf4 at the G1-to-S phase transition also blocks the phosphorylation of Mcm2 at this defined point of the cell cycle. -

Three Dact Gene Family Members Are Expressed During Embryonic Development
Three Dact Gene Family Members are Expressed During Embryonic Development and in the Adult Brains of Mice Daniel A Fisher, Saul Kivimäe, Jun Hoshino, Rowena Suriben, Pierre -Marie Martin, Nichol Baxter, Benjamin NR Cheyette Department of Psychiatry & Gra duate Programs in Developmental Biology and Neuroscience, University of California, San Francisco, 94143-2611 Correspondence: Benjamin NR Cheyette [email protected] 415.476.7826 Running Title: Mouse Dact Gene Family Expression Key Words: mouse, Dpr, Frodo, Thyex, Dact, Wnt, Dvl, expression, embryo, brain Supported by: NIH: MH01750 K08; NARSAD Young Investigator Award; NAAR award #551. Abstract Members of the Dact protein family were initially identified through binding to Dishevelled (Dvl), a cytoplasmic protein central to Wnt signaling. During mouse development, Dact1 is detected in the presomitic mesoderm and somites during segmentation, in the limb bud mesenchyme and other mesoderm-derived tissues, and in the central nervous system (CNS). Dact2 expression is most prominent during organogenesis of the thymus, kidneys, and salivary glands, with much lower levels in the somites and in the developing CNS. Dact3, not previously described in any organism, is expressed in the ventral region of maturing somites, limb bud and branchial arch mesenchyme, and in the embryonic CNS; of the three paralogs it is the most highly expressed in the adult cerebral cortex. These data are consistent with studies in other vertebrates showing that Dact paralogs have distinct signaling and developmental roles, and suggest they may differentially contribute to postnatal brain physiology. Introduction Signaling downstream of secreted Wnt ligands is a conserved process in multicellular animals that plays important roles during development and, when misregulated, contributes to cancer and other diseases (Polakis, 2000; Moon et al., 2002). -

DNA Replication Stress Response Involving PLK1, CDC6, POLQ
DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, E. Leconte, V. Schilling, K. Gordien, L. Brouchet, Mb Delisle, et al. To cite this version: C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, et al.. DNA replica- tion stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis, Nature Publishing Group: Open Access Journals - Option C, 2012, 1, pp.e30. 10.1038/oncsis.2012.29. hal-00817701 HAL Id: hal-00817701 https://hal.archives-ouvertes.fr/hal-00817701 Submitted on 9 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License Citation: Oncogenesis (2012) 1, e30; doi:10.1038/oncsis.2012.29 & 2012 Macmillan Publishers Limited All rights reserved 2157-9024/12 www.nature.com/oncsis ORIGINAL ARTICLE DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C Allera-Moreau1,2,7, I Rouquette2,7, B Lepage3, N Oumouhou3, M Walschaerts4, E Leconte5, V Schilling1, K Gordien2, L Brouchet2, MB Delisle1,2, J Mazieres1,2, JS Hoffmann1, P Pasero6 and C Cazaux1 Lung cancer is the leading cause of cancer deaths worldwide. -

Characterization of Budding Yeast Orc6 : a Dimerization Domain Is
Characterization of the role of Orc6 in the cell cycle of the budding yeast Saccharomyces cerevisiae by Jeffrey W. Semple A thesis presented to the University of Waterloo in fulfilment of the thesis requirement for the degree of Doctor of Philosophy in Biology Waterloo, Ontario, Canada, 2006 © Jeffrey W. Semple 2006 I hereby declare that I am the sole author of this thesis. This is a true copy of the thesis, including any required finalrevisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Abstract The heterohexameric origin recognition complex (ORC) acts as a scaffold for the G1 phase assembly of pre-replicative complexes. Only the Orc1-5 subunits are required for origin binding in budding yeast, yet Orc6 is an essential protein for cell proliferation. In comparison to other eukaryotic Orc6 proteins, budding yeast Orc6 appears to be quite divergent. Two-hybrid analysis revealed that Orc6 only weakly interacts with other ORC subunits. In this assay Orc6 showed a strong ability to self-associate, although the significance of this dimerization or multimerization remains unclear. Imaging of Orc6- eYFP revealed a punctate sub-nuclear localization pattern throughout the cell cycle, representing the first visualization of replication foci in live budding yeast cells. Orc6 was not detected at the site of division between mother and daughter cells, in contrast to observations from metazoans. An essential role for Orc6 in DNA replication was identified by depleting the protein before and during G1 phase. Surprisingly, Orc6 was required for entry into S phase after pre-replicative complex formation, in contrast to what has been observed for other ORC subunits. -

"The Genecards Suite: from Gene Data Mining to Disease Genome Sequence Analyses". In: Current Protocols in Bioinformat
The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses Gil Stelzer,1,5 Naomi Rosen,1,5 Inbar Plaschkes,1,2 Shahar Zimmerman,1 Michal Twik,1 Simon Fishilevich,1 Tsippi Iny Stein,1 Ron Nudel,1 Iris Lieder,2 Yaron Mazor,2 Sergey Kaplan,2 Dvir Dahary,2,4 David Warshawsky,3 Yaron Guan-Golan,3 Asher Kohn,3 Noa Rappaport,1 Marilyn Safran,1 and Doron Lancet1,6 1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel 2LifeMap Sciences Ltd., Tel Aviv, Israel 3LifeMap Sciences Inc., Marshfield, Massachusetts 4Toldot Genetics Ltd., Hod Hasharon, Israel 5These authors contributed equally to the paper 6Corresponding author GeneCards, the human gene compendium, enables researchers to effectively navigate and inter-relate the wide universe of human genes, diseases, variants, proteins, cells, and biological pathways. Our recently launched Version 4 has a revamped infrastructure facilitating faster data updates, better-targeted data queries, and friendlier user experience. It also provides a stronger foundation for the GeneCards suite of companion databases and analysis tools. Improved data unification includes gene-disease links via MalaCards and merged biological pathways via PathCards, as well as drug information and proteome expression. VarElect, another suite member, is a phenotype prioritizer for next-generation sequencing, leveraging the GeneCards and MalaCards knowledgebase. It au- tomatically infers direct and indirect scored associations between hundreds or even thousands of variant-containing genes and disease phenotype terms. Var- Elect’s capabilities, either independently or within TGex, our comprehensive variant analysis pipeline, help prepare for the challenge of clinical projects that involve thousands of exome/genome NGS analyses. -
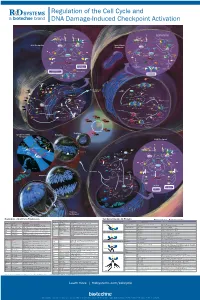
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

The Role of Dbf4-Dependent Protein Kinase in DNA Polymerase Ζ
INVESTIGATION The Role of Dbf4-Dependent Protein Kinase in DNA Polymerase z-Dependent Mutagenesis in Saccharomyces cerevisiae Luis N. Brandão,*,1,2 Rebecca Ferguson,1,* Irma Santoro,†,3 Sue Jinks-Robertson,†,‡ and Robert A. Sclafani* *Department of Biochemistry and Molecular Genetics, University of Colorado School of Medicine, Aurora, Colorado 80045, †Department of Biology, Emory University, Atlanta, Georgia 30322, and ‡Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina 27710 ABSTRACT The yeast Dbf4-dependent kinase (DDK) (composed of Dbf4 and Cdc7 subunits) is an essential, conserved Ser/Thr protein kinase that regulates multiple processes in the cell, including DNA replication, recombination and induced mutagenesis. Only DDK substrates important for replication and recombination have been identified. Consequently, the mechanism by which DDK regulates mutagenesis is unknown. The yeast mcm5-bob1 mutation that bypasses DDK’s essential role in DNA replication was used here to examine whether loss of DDK affects spontaneous as well as induced mutagenesis. Using the sensitive lys2DA746 frameshift reversion assay, we show DDK is required to generate “complex” spontaneous mutations, which are a hallmark of the Polz translesion synthesis DNA polymerase. DDK co-immunoprecipitated with the Rev7 regulatory, but not with the Rev3 polymerase subunit of Polz. Conversely, Rev7 bound mainly to the Cdc7 kinase subunit and not to Dbf4.TheRev7 subunit of Polz may be regulated by DDK phosphorylation as immunoprecipitates of yeast Cdc7 and also recombinant Xenopus DDK phosphorylated GST-Rev7 in vitro. In addition to promoting Polz- dependent mutagenesis, DDK was also important for generating Polz-independent large deletions that revert the lys2DA746 allele. -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

Anti-DBF4B (Aa 1-50) Polyclonal Antibody (DPABH- 12596) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-DBF4B (aa 1-50) polyclonal antibody (DPABH- 12596) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Antigen Description Regulatory subunit for CDC7 which activates its kinase activity thereby playing a central role in DNA replication and cell proliferation. Required for progression of S and M phases. The complex CDC7-DBF4B selectively phosphorylates MCM2 subunit at Ser-40 and then is involved in regulating the initiation of DNA replication during cell cycle. Immunogen Synthetic peptide within Human DBF4B aa 1-50 (N terminal). The exact sequence is proprietary. (NP_079380.2)Sequence: MSEPGKGDDCLELESSMAESRLRAPDLGVSRCLGKCQKNSPGARKHPFSG Database link: Q8NFT6-2 Run BLAST with Run BLAST with Isotype IgG Source/Host Rabbit Species Reactivity Human Purification Immunogen affinity purified Conjugate Unconjugated Applications WB Format Liquid Size 50 μg Preservative None Storage Shipped at 4°C. Store at 4°C short term (1-2 weeks). Upon delivery aliquot. Store at -20°C long term. Avoid freeze / thaw cycle. GENE INFORMATION Gene Name DBF4B DBF4 homolog B (S. cerevisiae) [ Homo sapiens ] Official Symbol DBF4B 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Synonyms DBF4B; DBF4 homolog B (S. cerevisiae); protein DBF4 homolog B; ASKL1; chifb; chiffon homolog B (Drosophila); DRF1; FLJ13087; ZDBF1B; zinc finger; DBF type containing 1B; chiffon homolog B; ASK-like protein 1; Dbf4-related factor 1; zinc finger, DBF-type containing 1B; activator of S-phase kinase-like protein 1; CHIFB; MGC15009; Entrez Gene ID 80174 Protein Refseq NP_079380 UniProt ID Q8NFT6 Chromosome Location 17q21.31 Function metal ion binding; nucleic acid binding; zinc ion binding; 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 2 © Creative Diagnostics All Rights Reserved. -

Crystal Structure of Rad53 Bound to Dbf4 and Cdc7 Received: 27 July 2016 Ahmad W
www.nature.com/scientificreports OPEN ‘AND’ logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7 Received: 27 July 2016 Ahmad W. Almawi1, Lindsay A. Matthews1,†, Larasati2, Polina Myrox2, Stephen Boulton3, Accepted: 07 September 2016 Christine Lai4, Trevor Moraes4, Giuseppe Melacini3, Rodolfo Ghirlando5, Bernard P. Duncker2 & Published: 29 September 2016 Alba Guarné1 Forkhead-associated (FHA) domains are phosphopeptide recognition modules found in many signaling proteins. The Saccharomyces cerevisiae protein kinase Rad53 is a key regulator of the DNA damage checkpoint and uses its two FHA domains to interact with multiple binding partners during the checkpoint response. One of these binding partners is the Dbf4-dependent kinase (DDK), a heterodimer composed of the Cdc7 kinase and its regulatory subunit Dbf4. Binding of Rad53 to DDK, through its N-terminal FHA (FHA1) domain, ultimately inhibits DDK kinase activity, thereby preventing firing of late origins. We have previously found that the FHA1 domain of Rad53 binds simultaneously to Dbf4 and a phosphoepitope, suggesting that this domain functions as an ‘AND’ logic gate. Here, we present the crystal structures of the FHA1 domain of Rad53 bound to Dbf4, in the presence and absence of a Cdc7 phosphorylated peptide. Our results reveal how the FHA1 uses a canonical binding interface to recognize the Cdc7 phosphopeptide and a non-canonical interface to bind Dbf4. Based on these data we propose a mechanism to explain how Rad53 enhances the specificity of FHA1-mediated transient interactions. Stress generated during DNA replication is one of the biggest hurdles proliferating cells face to preserve genome integrity. Therefore, eukaryotic cells have conserved surveillance mechanisms, known as cell cycle checkpoints, to detect and repair damage generated during DNA replication1–4. -

Downregulation of SNRPG Induces Cell Cycle Arrest and Sensitizes Human Glioblastoma Cells to Temozolomide by Targeting Myc Through a P53-Dependent Signaling Pathway
Cancer Biol Med 2020. doi: 10.20892/j.issn.2095-3941.2019.0164 ORIGINAL ARTICLE Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway Yulong Lan1,2*, Jiacheng Lou2*, Jiliang Hu1, Zhikuan Yu1, Wen Lyu1, Bo Zhang1,2 1Department of Neurosurgery, Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen 518020, China;2 Department of Neurosurgery, The Second Affiliated Hospital of Dalian Medical University, Dalian 116023, China ABSTRACT Objective: Temozolomide (TMZ) is commonly used for glioblastoma multiforme (GBM) chemotherapy. However, drug resistance limits its therapeutic effect in GBM treatment. RNA-binding proteins (RBPs) have vital roles in posttranscriptional events. While disturbance of RBP-RNA network activity is potentially associated with cancer development, the precise mechanisms are not fully known. The SNRPG gene, encoding small nuclear ribonucleoprotein polypeptide G, was recently found to be related to cancer incidence, but its exact function has yet to be elucidated. Methods: SNRPG knockdown was achieved via short hairpin RNAs. Gene expression profiling and Western blot analyses were used to identify potential glioma cell growth signaling pathways affected by SNRPG. Xenograft tumors were examined to determine the carcinogenic effects of SNRPG on glioma tissues. Results: The SNRPG-mediated inhibitory effect on glioma cells might be due to the targeted prevention of Myc and p53. In addition, the effects of SNRPG loss on p53 levels and cell cycle progression were found to be Myc-dependent. Furthermore, SNRPG was increased in TMZ-resistant GBM cells, and downregulation of SNRPG potentially sensitized resistant cells to TMZ, suggesting that SNRPG deficiency decreases the chemoresistance of GBM cells to TMZ via the p53 signaling pathway. -

A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes
A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes Deanna Petrochilos A dissertation presented in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2013 Reading Committee: Neil Abernethy, Chair John Gennari, Ali Shojaie Program Authorized to Offer Degree: Biomedical Informatics and Health Education UMI Number: 3588836 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. UMI 3588836 Published by ProQuest LLC (2013). Copyright in the Dissertation held by the Author. Microform Edition © ProQuest LLC. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, MI 48106 - 1346 ©Copyright 2013 Deanna Petrochilos University of Washington Abstract Using Graph-Based Methods to Integrate and Analyze Cancer Genomic Data Deanna Petrochilos Chair of the Supervisory Committee: Assistant Professor Neil Abernethy Biomedical Informatics and Health Education Studies of the genetic basis of complex disease present statistical and methodological challenges in the discovery of reliable and high-confidence genes that reveal biological phenomena underlying the etiology of disease or gene signatures prognostic of disease outcomes. This dissertation examines the capacity of graph-theoretical methods to model and analyze genomic information and thus facilitate using prior knowledge to create a more discrete and functionally relevant feature space.