The Characterization of Dbf4 Interactions and Roles in Genome Replication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mcm2 Is a Target of Regulation by Cdc7–Dbf4 During the Initiation of DNA Synthesis
Downloaded from genesdev.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis Ming Lei,1 Yasuo Kawasaki,1,2 Michael R. Young,1,3 Makoto Kihara,2 Akio Sugino,2 and Bik K. Tye1,4 1Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, New York 14853-2703 USA; 2Research Institute for Microbial Diseases, Osaka University, Osaka 565, Japan The initiation of DNA synthesis is an important cell cycle event that defines the beginning of S phase. This critical event involves the participation of proteins whose functions are regulated by cyclin dependent protein kinases (Cdks). The Mcm2–7 proteins are a family of six conserved proteins that are essential for the initiation of DNA synthesis in all eukaryotes. In Saccharomyces cerevisiae, members of the Mcm2–7 family undergo cell cycle-specific phosphorylation. Phosphorylation of Mcm proteins at the beginning of S phase coincides with the removal of these proteins from chromatin and the onset of DNA synthesis. In this study, we identified DBF4, which encodes the regulatory subunit of a Cdk-like protein kinase Cdc7–Dbf4, in a screen for second site suppressors of mcm2-1. The dbf4 suppressor mutation restores competence to initiate DNA synthesis to the mcm2-1 mutant. Cdc7–Dbf4 interacts physically with Mcm2 and phosphorylates Mcm2 and three other members of the Mcm2–7 family in vitro. Blocking the kinase activity of Cdc7–Dbf4 at the G1-to-S phase transition also blocks the phosphorylation of Mcm2 at this defined point of the cell cycle. -

Three Dact Gene Family Members Are Expressed During Embryonic Development
Three Dact Gene Family Members are Expressed During Embryonic Development and in the Adult Brains of Mice Daniel A Fisher, Saul Kivimäe, Jun Hoshino, Rowena Suriben, Pierre -Marie Martin, Nichol Baxter, Benjamin NR Cheyette Department of Psychiatry & Gra duate Programs in Developmental Biology and Neuroscience, University of California, San Francisco, 94143-2611 Correspondence: Benjamin NR Cheyette [email protected] 415.476.7826 Running Title: Mouse Dact Gene Family Expression Key Words: mouse, Dpr, Frodo, Thyex, Dact, Wnt, Dvl, expression, embryo, brain Supported by: NIH: MH01750 K08; NARSAD Young Investigator Award; NAAR award #551. Abstract Members of the Dact protein family were initially identified through binding to Dishevelled (Dvl), a cytoplasmic protein central to Wnt signaling. During mouse development, Dact1 is detected in the presomitic mesoderm and somites during segmentation, in the limb bud mesenchyme and other mesoderm-derived tissues, and in the central nervous system (CNS). Dact2 expression is most prominent during organogenesis of the thymus, kidneys, and salivary glands, with much lower levels in the somites and in the developing CNS. Dact3, not previously described in any organism, is expressed in the ventral region of maturing somites, limb bud and branchial arch mesenchyme, and in the embryonic CNS; of the three paralogs it is the most highly expressed in the adult cerebral cortex. These data are consistent with studies in other vertebrates showing that Dact paralogs have distinct signaling and developmental roles, and suggest they may differentially contribute to postnatal brain physiology. Introduction Signaling downstream of secreted Wnt ligands is a conserved process in multicellular animals that plays important roles during development and, when misregulated, contributes to cancer and other diseases (Polakis, 2000; Moon et al., 2002). -

DNA: the Timeline and Evidence of Discovery
1/19/2017 DNA: The Timeline and Evidence of Discovery Interactive Click and Learn (Ann Brokaw Rocky River High School) Introduction For almost a century, many scientists paved the way to the ultimate discovery of DNA and its double helix structure. Without the work of these pioneering scientists, Watson and Crick may never have made their ground-breaking double helix model, published in 1953. The knowledge of how genetic material is stored and copied in this molecule gave rise to a new way of looking at and manipulating biological processes, called molecular biology. The breakthrough changed the face of biology and our lives forever. Watch The Double Helix short film (approximately 15 minutes) – hyperlinked here. 1 1/19/2017 1865 The Garden Pea 1865 The Garden Pea In 1865, Gregor Mendel established the foundation of genetics by unraveling the basic principles of heredity, though his work would not be recognized as “revolutionary” until after his death. By studying the common garden pea plant, Mendel demonstrated the inheritance of “discrete units” and introduced the idea that the inheritance of these units from generation to generation follows particular patterns. These patterns are now referred to as the “Laws of Mendelian Inheritance.” 2 1/19/2017 1869 The Isolation of “Nuclein” 1869 Isolated Nuclein Friedrich Miescher, a Swiss researcher, noticed an unknown precipitate in his work with white blood cells. Upon isolating the material, he noted that it resisted protein-digesting enzymes. Why is it important that the material was not digested by the enzymes? Further work led him to the discovery that the substance contained carbon, hydrogen, nitrogen and large amounts of phosphorus with no sulfur. -

DNA Replication Stress Response Involving PLK1, CDC6, POLQ
DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, E. Leconte, V. Schilling, K. Gordien, L. Brouchet, Mb Delisle, et al. To cite this version: C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, et al.. DNA replica- tion stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis, Nature Publishing Group: Open Access Journals - Option C, 2012, 1, pp.e30. 10.1038/oncsis.2012.29. hal-00817701 HAL Id: hal-00817701 https://hal.archives-ouvertes.fr/hal-00817701 Submitted on 9 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License Citation: Oncogenesis (2012) 1, e30; doi:10.1038/oncsis.2012.29 & 2012 Macmillan Publishers Limited All rights reserved 2157-9024/12 www.nature.com/oncsis ORIGINAL ARTICLE DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C Allera-Moreau1,2,7, I Rouquette2,7, B Lepage3, N Oumouhou3, M Walschaerts4, E Leconte5, V Schilling1, K Gordien2, L Brouchet2, MB Delisle1,2, J Mazieres1,2, JS Hoffmann1, P Pasero6 and C Cazaux1 Lung cancer is the leading cause of cancer deaths worldwide. -
![Photograph 51, by Rosalind Franklin (1952) [1]](https://docslib.b-cdn.net/cover/5767/photograph-51-by-rosalind-franklin-1952-1-745767.webp)
Photograph 51, by Rosalind Franklin (1952) [1]
Published on The Embryo Project Encyclopedia (https://embryo.asu.edu) Photograph 51, by Rosalind Franklin (1952) [1] By: Hernandez, Victoria Keywords: X-ray crystallography [2] DNA [3] DNA Helix [4] On 6 May 1952, at King´s College London in London, England, Rosalind Franklin photographed her fifty-first X-ray diffraction pattern of deoxyribosenucleic acid, or DNA. Photograph 51, or Photo 51, revealed information about DNA´s three-dimensional structure by displaying the way a beam of X-rays scattered off a pure fiber of DNA. Franklin took Photo 51 after scientists confirmed that DNA contained genes [5]. Maurice Wilkins, Franklin´s colleague showed James Watson [6] and Francis Crick [7] Photo 51 without Franklin´s knowledge. Watson and Crick used that image to develop their structural model of DNA. In 1962, after Franklin´s death, Watson, Crick, and Wilkins shared the Nobel Prize in Physiology or Medicine [8] for their findings about DNA. Franklin´s Photo 51 helped scientists learn more about the three-dimensional structure of DNA and enabled scientists to understand DNA´s role in heredity. X-ray crystallography, the technique Franklin used to produce Photo 51 of DNA, is a method scientists use to determine the three-dimensional structure of a crystal. Crystals are solids with regular, repeating units of atoms. Some biological macromolecules, such as DNA, can form fibers suitable for analysis using X-ray crystallography because their solid forms consist of atoms arranged in a regular pattern. Photo 51 used DNA fibers, DNA crystals first produced in the 1970s. To perform an X-ray crystallography, scientists mount a purified fiber or crystal in an X-ray tube. -

Characterization of Budding Yeast Orc6 : a Dimerization Domain Is
Characterization of the role of Orc6 in the cell cycle of the budding yeast Saccharomyces cerevisiae by Jeffrey W. Semple A thesis presented to the University of Waterloo in fulfilment of the thesis requirement for the degree of Doctor of Philosophy in Biology Waterloo, Ontario, Canada, 2006 © Jeffrey W. Semple 2006 I hereby declare that I am the sole author of this thesis. This is a true copy of the thesis, including any required finalrevisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Abstract The heterohexameric origin recognition complex (ORC) acts as a scaffold for the G1 phase assembly of pre-replicative complexes. Only the Orc1-5 subunits are required for origin binding in budding yeast, yet Orc6 is an essential protein for cell proliferation. In comparison to other eukaryotic Orc6 proteins, budding yeast Orc6 appears to be quite divergent. Two-hybrid analysis revealed that Orc6 only weakly interacts with other ORC subunits. In this assay Orc6 showed a strong ability to self-associate, although the significance of this dimerization or multimerization remains unclear. Imaging of Orc6- eYFP revealed a punctate sub-nuclear localization pattern throughout the cell cycle, representing the first visualization of replication foci in live budding yeast cells. Orc6 was not detected at the site of division between mother and daughter cells, in contrast to observations from metazoans. An essential role for Orc6 in DNA replication was identified by depleting the protein before and during G1 phase. Surprisingly, Orc6 was required for entry into S phase after pre-replicative complex formation, in contrast to what has been observed for other ORC subunits. -

A Brief History of Genetics
A Brief History of Genetics A Brief History of Genetics By Chris Rider A Brief History of Genetics By Chris Rider This book first published 2020 Cambridge Scholars Publishing Lady Stephenson Library, Newcastle upon Tyne, NE6 2PA, UK British Library Cataloguing in Publication Data A catalogue record for this book is available from the British Library Copyright © 2020 by Chris Rider All rights for this book reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without the prior permission of the copyright owner. ISBN (10): 1-5275-5885-1 ISBN (13): 978-1-5275-5885-4 Cover A cartoon of the double-stranded helix structure of DNA overlies the sequence of the gene encoding the A protein chain of human haemoglobin. Top left is a portrait of Gregor Mendel, the founding father of genetics, and bottom right is a portrait of Thomas Hunt Morgan, the first winner of a Nobel Prize for genetics. To my wife for her many years of love, support, patience and sound advice TABLE OF CONTENTS List of Figures.......................................................................................... viii List of Text Boxes and Tables .................................................................... x Foreword .................................................................................................. xi Acknowledgements ................................................................................. xiii Chapter 1 ................................................................................................... -

"The Genecards Suite: from Gene Data Mining to Disease Genome Sequence Analyses". In: Current Protocols in Bioinformat
The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses Gil Stelzer,1,5 Naomi Rosen,1,5 Inbar Plaschkes,1,2 Shahar Zimmerman,1 Michal Twik,1 Simon Fishilevich,1 Tsippi Iny Stein,1 Ron Nudel,1 Iris Lieder,2 Yaron Mazor,2 Sergey Kaplan,2 Dvir Dahary,2,4 David Warshawsky,3 Yaron Guan-Golan,3 Asher Kohn,3 Noa Rappaport,1 Marilyn Safran,1 and Doron Lancet1,6 1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel 2LifeMap Sciences Ltd., Tel Aviv, Israel 3LifeMap Sciences Inc., Marshfield, Massachusetts 4Toldot Genetics Ltd., Hod Hasharon, Israel 5These authors contributed equally to the paper 6Corresponding author GeneCards, the human gene compendium, enables researchers to effectively navigate and inter-relate the wide universe of human genes, diseases, variants, proteins, cells, and biological pathways. Our recently launched Version 4 has a revamped infrastructure facilitating faster data updates, better-targeted data queries, and friendlier user experience. It also provides a stronger foundation for the GeneCards suite of companion databases and analysis tools. Improved data unification includes gene-disease links via MalaCards and merged biological pathways via PathCards, as well as drug information and proteome expression. VarElect, another suite member, is a phenotype prioritizer for next-generation sequencing, leveraging the GeneCards and MalaCards knowledgebase. It au- tomatically infers direct and indirect scored associations between hundreds or even thousands of variant-containing genes and disease phenotype terms. Var- Elect’s capabilities, either independently or within TGex, our comprehensive variant analysis pipeline, help prepare for the challenge of clinical projects that involve thousands of exome/genome NGS analyses. -

STS.003 the Rise of Modern Science Spring 2008
MIT OpenCourseWare http://ocw.mit.edu STS.003 The Rise of Modern Science Spring 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. STS.003 Spring 2007 Keywords for Week 12 Lecture 20: Eugenics Darwin, Descent of Man (1871) Francis Galton, “eugenics” (1883) “the science of improving the stock” Jean Baptiste Lamarck Inheritance of Acquired Characteristics Richard Dugdale, The Jukes: A Study in Crime, Pauperism, Disease and Heredity (1874) Karl Pearson Galton Laboratory for National Eugenics Charles Davenport Cold Spring Harbor Eugenics Record Office Arthur Estabrook, The Jukes in 1915 (1916) Henry Goddard, The Kallikak Family: A Study in the Heredity of Feeble-mindedness (1912) Deborah Kallikak Race Suicide Fitter Family Contests Sterilization Laws Buck v. Bell, 1927 Carrie Buck Justice Oliver Wendell Holmes, Jr. “Three generations of imbeciles is enough” Quotes But are not our physical faculties and the strength, dexterity and acuteness of our senses, to be numbered among the qualities whose perfection in the individual may be transmitted? Observation of the various breeds of domestic animals inclines us to believe that they are, and we can confirm this by direct observation of the human race. Condorcet, Future Progress of the Human Spirit (1795) If a twentieth part of the cost and pains were spent in measures for the improvement of the human race that is spent on the improvement of the breed of horses and cattle, what a galaxy of genius we might create. Francis Galton, “Hereditary Character and Talent” (1864) Both sexes ought to refrain from marriage if they are in any marked degree inferior in body or mind but such hopes are Utopian and will never be even partially realized until the laws of inheritance are thoroughly known. -

Video Worksheet - Secret of Photo 51
Name __________________________________________ Period ________ Date _______________ Video Worksheet - Secret of Photo 51 We are watching the NOVA video entitled “Secret of Photo 51.” This video demonstrates the race to determine the structure of DNA during the 1940s and 1950s. Of particular interest through the video, we see how a female scientist, Rosalind Franklin, was of essential to the discovery. 1. In 1962, who was awarded the Nobel Prize for the discovery of the structure of DNA? (Please give three names) 2. Which of those three wrote the book “The Double Helix”? 3. How did he characterize Rosalind Franklin in his book? 4. What was Rosalind Franklin like as a child? 5. Where did she study physics and chemistry? 6. What city did Rosalind Franklin perfect her work in crystallography in? 7. Where in England is Rosalind Franklin offered a position? 8. What misunderstanding occurred between Franklin and Maurice Wilkins? 9. What was the environment like for a female scientist? What nicknames was Franklin given? 10. What two forms of DNA did Franklin discover? Name __________________________________________ Period ________ Date _______________ 11. Who is in the audience listening to Franklin’s talk? What does he want to do with her information? 12. Who gives away Franklin’s unpublished work? To whom does he give it to? 13. When did Franklin get her best picture? What did she title it? 14. The race is now in earnest. How do the “discoverers” come to their conclusion about the structure of DNA? What information did they need? 15. Did Franklin approve of the model in 1953? 16. -
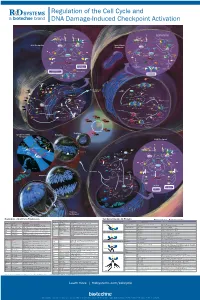
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

The Role of Dbf4-Dependent Protein Kinase in DNA Polymerase Ζ
INVESTIGATION The Role of Dbf4-Dependent Protein Kinase in DNA Polymerase z-Dependent Mutagenesis in Saccharomyces cerevisiae Luis N. Brandão,*,1,2 Rebecca Ferguson,1,* Irma Santoro,†,3 Sue Jinks-Robertson,†,‡ and Robert A. Sclafani* *Department of Biochemistry and Molecular Genetics, University of Colorado School of Medicine, Aurora, Colorado 80045, †Department of Biology, Emory University, Atlanta, Georgia 30322, and ‡Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina 27710 ABSTRACT The yeast Dbf4-dependent kinase (DDK) (composed of Dbf4 and Cdc7 subunits) is an essential, conserved Ser/Thr protein kinase that regulates multiple processes in the cell, including DNA replication, recombination and induced mutagenesis. Only DDK substrates important for replication and recombination have been identified. Consequently, the mechanism by which DDK regulates mutagenesis is unknown. The yeast mcm5-bob1 mutation that bypasses DDK’s essential role in DNA replication was used here to examine whether loss of DDK affects spontaneous as well as induced mutagenesis. Using the sensitive lys2DA746 frameshift reversion assay, we show DDK is required to generate “complex” spontaneous mutations, which are a hallmark of the Polz translesion synthesis DNA polymerase. DDK co-immunoprecipitated with the Rev7 regulatory, but not with the Rev3 polymerase subunit of Polz. Conversely, Rev7 bound mainly to the Cdc7 kinase subunit and not to Dbf4.TheRev7 subunit of Polz may be regulated by DDK phosphorylation as immunoprecipitates of yeast Cdc7 and also recombinant Xenopus DDK phosphorylated GST-Rev7 in vitro. In addition to promoting Polz- dependent mutagenesis, DDK was also important for generating Polz-independent large deletions that revert the lys2DA746 allele.