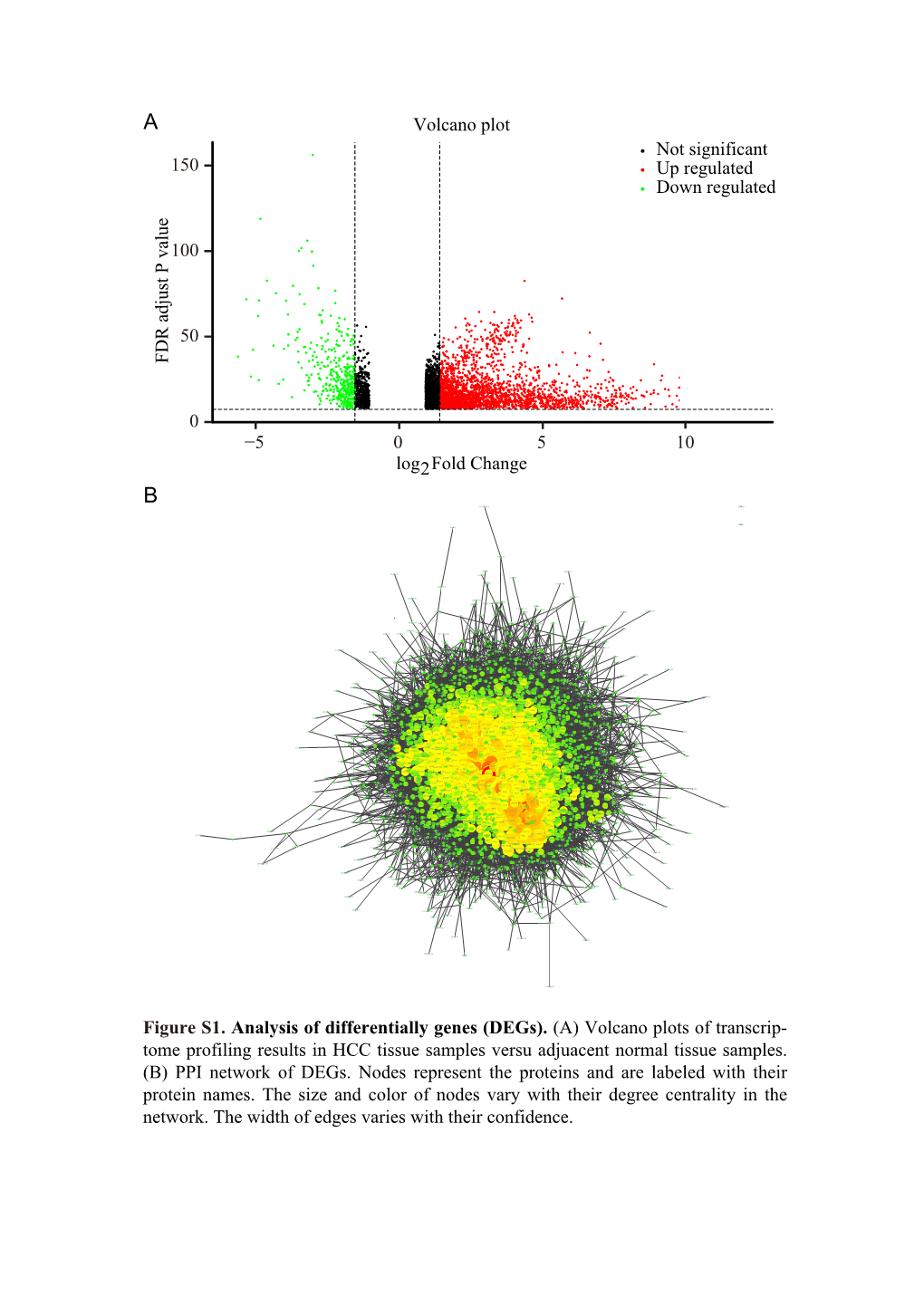
5 0 5 10 Log2 Fold Change FDR Adjust P Value Volcano Plot 0 50 100 150
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Associations Between Voltage-Gated Calcium Channels (Vgccs) and Autism Spectrum Disorder (ASD)
Liao and Li Molecular Brain (2020) 13:96 https://doi.org/10.1186/s13041-020-00634-0 REVIEW Open Access Genetic associations between voltage- gated calcium channels and autism spectrum disorder: a systematic review Xiaoli Liao1,2 and Yamin Li2* Abstract Objectives: The present review systematically summarized existing publications regarding the genetic associations between voltage-gated calcium channels (VGCCs) and autism spectrum disorder (ASD). Methods: A comprehensive literature search was conducted to gather pertinent studies in three online databases. Two authors independently screened the included records based on the selection criteria. Discrepancies in each step were settled through discussions. Results: From 1163 resulting searched articles, 28 were identified for inclusion. The most prominent among the VGCCs variants found in ASD were those falling within loci encoding the α subunits, CACNA1A, CACNA1B, CACN A1C, CACNA1D, CACNA1E, CACNA1F, CACNA1G, CACNA1H, and CACNA1I as well as those of their accessory subunits CACNB2, CACNA2D3, and CACNA2D4. Two signaling pathways, the IP3-Ca2+ pathway and the MAPK pathway, were identified as scaffolds that united genetic lesions into a consensus etiology of ASD. Conclusions: Evidence generated from this review supports the role of VGCC genetic variants in the pathogenesis of ASD, making it a promising therapeutic target. Future research should focus on the specific mechanism that connects VGCC genetic variants to the complex ASD phenotype. Keywords: Autism spectrum disorder, Voltage-gated calcium -

Supplemental Figure 1. Vimentin
Double mutant specific genes Transcript gene_assignment Gene Symbol RefSeq FDR Fold- FDR Fold- FDR Fold- ID (single vs. Change (double Change (double Change wt) (single vs. wt) (double vs. single) (double vs. wt) vs. wt) vs. single) 10485013 BC085239 // 1110051M20Rik // RIKEN cDNA 1110051M20 gene // 2 E1 // 228356 /// NM 1110051M20Ri BC085239 0.164013 -1.38517 0.0345128 -2.24228 0.154535 -1.61877 k 10358717 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 /// BC 1700025G04Rik NM_197990 0.142593 -1.37878 0.0212926 -3.13385 0.093068 -2.27291 10358713 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 1700025G04Rik NM_197990 0.0655213 -1.71563 0.0222468 -2.32498 0.166843 -1.35517 10481312 NM_027283 // 1700026L06Rik // RIKEN cDNA 1700026L06 gene // 2 A3 // 69987 /// EN 1700026L06Rik NM_027283 0.0503754 -1.46385 0.0140999 -2.19537 0.0825609 -1.49972 10351465 BC150846 // 1700084C01Rik // RIKEN cDNA 1700084C01 gene // 1 H3 // 78465 /// NM_ 1700084C01Rik BC150846 0.107391 -1.5916 0.0385418 -2.05801 0.295457 -1.29305 10569654 AK007416 // 1810010D01Rik // RIKEN cDNA 1810010D01 gene // 7 F5 // 381935 /// XR 1810010D01Rik AK007416 0.145576 1.69432 0.0476957 2.51662 0.288571 1.48533 10508883 NM_001083916 // 1810019J16Rik // RIKEN cDNA 1810019J16 gene // 4 D2.3 // 69073 / 1810019J16Rik NM_001083916 0.0533206 1.57139 0.0145433 2.56417 0.0836674 1.63179 10585282 ENSMUST00000050829 // 2010007H06Rik // RIKEN cDNA 2010007H06 gene // --- // 6984 2010007H06Rik ENSMUST00000050829 0.129914 -1.71998 0.0434862 -2.51672 -

The Mineralocorticoid Receptor Leads to Increased Expression of EGFR
www.nature.com/scientificreports OPEN The mineralocorticoid receptor leads to increased expression of EGFR and T‑type calcium channels that support HL‑1 cell hypertrophy Katharina Stroedecke1,2, Sandra Meinel1,2, Fritz Markwardt1, Udo Kloeckner1, Nicole Straetz1, Katja Quarch1, Barbara Schreier1, Michael Kopf1, Michael Gekle1 & Claudia Grossmann1* The EGF receptor (EGFR) has been extensively studied in tumor biology and recently a role in cardiovascular pathophysiology was suggested. The mineralocorticoid receptor (MR) is an important efector of the renin–angiotensin–aldosterone‑system and elicits pathophysiological efects in the cardiovascular system; however, the underlying molecular mechanisms are unclear. Our aim was to investigate the importance of EGFR for MR‑mediated cardiovascular pathophysiology because MR is known to induce EGFR expression. We identifed a SNP within the EGFR promoter that modulates MR‑induced EGFR expression. In RNA‑sequencing and qPCR experiments in heart tissue of EGFR KO and WT mice, changes in EGFR abundance led to diferential expression of cardiac ion channels, especially of the T‑type calcium channel CACNA1H. Accordingly, CACNA1H expression was increased in WT mice after in vivo MR activation by aldosterone but not in respective EGFR KO mice. Aldosterone‑ and EGF‑responsiveness of CACNA1H expression was confrmed in HL‑1 cells by Western blot and by measuring peak current density of T‑type calcium channels. Aldosterone‑induced CACNA1H protein expression could be abrogated by the EGFR inhibitor AG1478. Furthermore, inhibition of T‑type calcium channels with mibefradil or ML218 reduced diameter, volume and BNP levels in HL‑1 cells. In conclusion the MR regulates EGFR and CACNA1H expression, which has an efect on HL‑1 cell diameter, and the extent of this regulation seems to depend on the SNP‑216 (G/T) genotype. -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Supplementary Table S1. Upregulated Genes Differentially
Supplementary Table S1. Upregulated genes differentially expressed in athletes (p < 0.05 and 1.3-fold change) Gene Symbol p Value Fold Change 221051_s_at NMRK2 0.01 2.38 236518_at CCDC183 0.00 2.05 218804_at ANO1 0.00 2.05 234675_x_at 0.01 2.02 207076_s_at ASS1 0.00 1.85 209135_at ASPH 0.02 1.81 228434_at BTNL9 0.03 1.81 229985_at BTNL9 0.01 1.79 215795_at MYH7B 0.01 1.78 217979_at TSPAN13 0.01 1.77 230992_at BTNL9 0.01 1.75 226884_at LRRN1 0.03 1.74 220039_s_at CDKAL1 0.01 1.73 236520_at 0.02 1.72 219895_at TMEM255A 0.04 1.72 201030_x_at LDHB 0.00 1.69 233824_at 0.00 1.69 232257_s_at 0.05 1.67 236359_at SCN4B 0.04 1.64 242868_at 0.00 1.63 1557286_at 0.01 1.63 202780_at OXCT1 0.01 1.63 1556542_a_at 0.04 1.63 209992_at PFKFB2 0.04 1.63 205247_at NOTCH4 0.01 1.62 1554182_at TRIM73///TRIM74 0.00 1.61 232892_at MIR1-1HG 0.02 1.61 204726_at CDH13 0.01 1.6 1561167_at 0.01 1.6 1565821_at 0.01 1.6 210169_at SEC14L5 0.01 1.6 236963_at 0.02 1.6 1552880_at SEC16B 0.02 1.6 235228_at CCDC85A 0.02 1.6 1568623_a_at SLC35E4 0.00 1.59 204844_at ENPEP 0.00 1.59 1552256_a_at SCARB1 0.02 1.59 1557283_a_at ZNF519 0.02 1.59 1557293_at LINC00969 0.03 1.59 231644_at 0.01 1.58 228115_at GAREM1 0.01 1.58 223687_s_at LY6K 0.02 1.58 231779_at IRAK2 0.03 1.58 243332_at LOC105379610 0.04 1.58 232118_at 0.01 1.57 203423_at RBP1 0.02 1.57 AMY1A///AMY1B///AMY1C///AMY2A///AMY2B// 208498_s_at 0.03 1.57 /AMYP1 237154_at LOC101930114 0.00 1.56 1559691_at 0.01 1.56 243481_at RHOJ 0.03 1.56 238834_at MYLK3 0.01 1.55 213438_at NFASC 0.02 1.55 242290_at TACC1 0.04 1.55 ANKRD20A1///ANKRD20A12P///ANKRD20A2/// -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genomic Comparison of Indigenous African and Northern European
Animal Science Publications Animal Science 5-2017 Genomic Comparison of Indigenous African and Northern European Chickens Reveals Putative Mechanisms of Stress Tolerance Related to Environmental Selection Pressure Damarius S. Fleming Iowa State University Steffen Weigend Friedrich-Loeffler-Institut Henner Simianer University of Göttingen Annett eiW gend Friedrich-Loeffler-Institut Follow this and additional works at: https://lib.dr.iastate.edu/ans_pubs MaxP aRrothschit of theldAgriculture Commons, Ecology and Evolutionary Biology Commons, Genetics and GeIowanomic State Usn Civommonersity, mfrsoth, asndc@i theastaPteoultr.edu y or Avian Science Commons TheSee nex tompc page forle addte bitioniblaiol agruthorapshic information for this item can be found at https://lib.dr.iastate.edu/ ans_pubs/368. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Article is brought to you for free and open access by the Animal Science at Iowa State University Digital Repository. It has been accepted for inclusion in Animal Science Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Genomic Comparison of Indigenous African and Northern European Chickens Reveals Putative Mechanisms of Stress Tolerance Related to Environmental Selection Pressure Abstract Global climate change is increasing the magnitude of environmental stressors, such as temperature, pathogens, and drought, that limit the survivability and sustainability of livestock production. Poultry production and its expansion is dependent upon robust animals that are able to cope with stressors in multiple environments. Understanding the genetic strategies that indigenous, noncommercial breeds have evolved to survive in their environment could help to elucidate molecular mechanisms underlying biological traits of environmental adaptation. -

Supplementary Figure S4
18DCIS 18IDC Supplementary FigureS4 22DCIS 22IDC C D B A E (0.77) (0.78) 16DCIS 14DCIS 28DCIS 16IDC 28IDC (0.43) (0.49) 0 ADAMTS12 (p.E1469K) 14IDC ERBB2, LASP1,CDK12( CCNE1 ( NUTM2B SDHC,FCGR2B,PBX1,TPR( CD1D, B4GALT3, BCL9, FLG,NUP21OL,TPM3,TDRD10,RIT1,LMNA,PRCC,NTRK1 0 ADAMTS16 (p.E67K) (0.67) (0.89) (0.54) 0 ARHGEF38 (p.P179Hfs*29) 0 ATG9B (p.P823S) (0.68) (1.0) ARID5B, CCDC6 CCNE1, TSHZ3,CEP89 CREB3L2,TRIM24 BRAF, EGFR (7p11); 0 ABRACL (p.R35H) 0 CATSPER1 (p.P152H) 0 ADAMTS18 (p.Y799C) 19q12 0 CCDC88C (p.X1371_splice) (0) 0 ADRA1A (p.P327L) (10q22.3) 0 CCNF (p.D637N) −4 −2 −4 −2 0 AKAP4 (p.G454A) 0 CDYL (p.Y353Lfs*5) −4 −2 Log2 Ratio Log2 Ratio −4 −2 Log2 Ratio Log2 Ratio 0 2 4 0 2 4 0 ARID2 (p.R1068H) 0 COL27A1 (p.G646E) 0 2 4 0 2 4 2 EDRF1 (p.E521K) 0 ARPP21 (p.P791L) ) 0 DDX11 (p.E78K) 2 GPR101, p.A174V 0 ARPP21 (p.P791T) 0 DMGDH (p.W606C) 5 ANP32B, p.G237S 16IDC (Ploidy:2.01) 16DCIS (Ploidy:2.02) 14IDC (Ploidy:2.01) 14DCIS (Ploidy:2.9) -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 Log Ratio Log Ratio Log Ratio Log Ratio 12DCIS 0 ASPM (p.S222T) Log Ratio Log Ratio 0 FMN2 (p.G941A) 20 1 2 3 2 0 1 2 3 2 ERBB3 (p.D297Y) 2 0 1 2 3 20 1 2 3 0 ATRX (p.L1276I) 20 1 2 3 2 0 1 2 3 0 GALNT18 (p.F92L) 2 MAPK4, p.H147Y 0 GALNTL6 (p.E236K) 5 C11orf1, p.Y53C (10q21.2); 0 ATRX (p.R1401W) PIK3CA, p.H1047R 28IDC (Ploidy:2.0) 28DCIS (Ploidy:2.0) 22IDC (Ploidy:3.7) 22DCIS (Ploidy:4.1) 18IDC (Ploidy:3.9) 18DCIS (Ploidy:2.3) 17q12 0 HCFC1 (p.S2025C) 2 LCMT1 (p.S34A) 0 ATXN7L2 (p.X453_splice) SPEN, p.P677Lfs*13 CBFB 1 2 3 4 5 6 7 8 9 10 11 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

(SLC3A2) Sustains Amino Acid and Nucleotide Availability for Cell Cycle
www.nature.com/scientificreports OPEN CD98hc (SLC3A2) sustains amino acid and nucleotide availability for cell cycle progression Received: 16 April 2019 Sara Cano-Crespo1, Josep Chillarón2, Alexandra Junza3,4, Gonzalo Fernández-Miranda1, Accepted: 13 September 2019 Judit García5,6, Christine Polte7, Laura R. de la Ballina 8,9, Zoya Ignatova7, Óscar Yanes 3,4, Published: xx xx xxxx Antonio Zorzano 1,4,10, Camille Stephan-Otto Attolini 1 & Manuel Palacín1,6,10 CD98 heavy chain (CD98hc) forms heteromeric amino acid (AA) transporters by interacting with diferent light chains. Cancer cells overexpress CD98hc-transporters in order to meet their increased nutritional and antioxidant demands, since they provide branched-chain AA (BCAA) and aromatic AA (AAA) availability while protecting cells from oxidative stress. Here we show that BCAA and AAA shortage phenocopies the inhibition of mTORC1 signalling, protein synthesis and cell proliferation caused by CD98hc ablation. Furthermore, our data indicate that CD98hc sustains glucose uptake and glycolysis, and, as a consequence, the pentose phosphate pathway (PPP). Thus, loss of CD98hc triggers a dramatic reduction in the nucleotide pool, which leads to replicative stress in these cells, as evidenced by the enhanced DNA Damage Response (DDR), S-phase delay and diminished rate of mitosis, all recovered by nucleoside supplementation. In addition, proper BCAA and AAA availability sustains the expression of the enzyme ribonucleotide reductase. In this regard, BCAA and AAA shortage results in decreased content of deoxynucleotides that triggers replicative stress, also recovered by nucleoside supplementation. On the basis of our fndings, we conclude that CD98hc plays a central role in AA and glucose cellular nutrition, redox homeostasis and nucleotide availability, all key for cell proliferation. -

Abnormal Spermatogenesis and Reduced Fertility in Transition Nuclear Protein 1-Deficient Mice
Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice Y. Eugene Yu*†,Yun Zhang*, Emmanual Unni*‡, Cynthia R. Shirley*, Jian M. Deng§, Lonnie D. Russell¶, Michael M. Weil*, Richard R. Behringer§, and Marvin L. Meistrich*ʈ Departments of *Experimental Radiation Oncology, and §Molecular Genetics, University of Texas M. D. Anderson Cancer Center, Houston, TX 77030-4095; and ¶Department of Physiology, Southern Illinois University, School of Medicine, Carbondale, IL 62901 Edited by Richard D. Palmiter, University of Washington School of Medicine, Seattle, WA, and approved February 22, 2000 (received for review May 3, 1999) Transition nuclear proteins (TPs), the major proteins found in (15), suggesting some functional relationship between the three chromatin of condensing spermatids, are believed to be important proteins exists. Tnp1, however, is on a separate chromosome and for histone displacement and chromatin condensation during is not clearly related to the other three proteins. mammalian spermatogenesis. We generated mice lacking the ma- In vitro, TP1 decreases the melting temperature of DNA (16) jor TP, TP1, by targeted deletion of the Tnp1 gene in mouse and relaxes the DNA in nucleosomal core particles (17), which embryonic stem cells. Surprisingly, testis weights and sperm pro- led to the proposal that TP1 reduces the interaction of DNA duction were normal in the mutant mice, and only subtle abnor- with the nucleosome core. In contrast, TP2 increases the malities were observed in sperm morphology. Electron microscopy melting temperature of DNA and compacts the DNA in revealed large rod-like structures in the chromatin of mutant step nucleosomal cores, suggesting that it is a DNA-condensing 13 spermatids, in contrast to the fine chromatin fibrils observed in protein (18). -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties.