Use of Species Distribution Modeling in the Deep Sea
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Meta-Ecosystems: a Theoretical Framework for a Spatial Ecosystem Ecology
Ecology Letters, (2003) 6: 673–679 doi: 10.1046/j.1461-0248.2003.00483.x IDEAS AND PERSPECTIVES Meta-ecosystems: a theoretical framework for a spatial ecosystem ecology Abstract Michel Loreau1*, Nicolas This contribution proposes the meta-ecosystem concept as a natural extension of the Mouquet2,4 and Robert D. Holt3 metapopulation and metacommunity concepts. A meta-ecosystem is defined as a set of 1Laboratoire d’Ecologie, UMR ecosystems connected by spatial flows of energy, materials and organisms across 7625, Ecole Normale Supe´rieure, ecosystem boundaries. This concept provides a powerful theoretical tool to understand 46 rue d’Ulm, F–75230 Paris the emergent properties that arise from spatial coupling of local ecosystems, such as Cedex 05, France global source–sink constraints, diversity–productivity patterns, stabilization of ecosystem 2Department of Biological processes and indirect interactions at landscape or regional scales. The meta-ecosystem Science and School of perspective thereby has the potential to integrate the perspectives of community and Computational Science and Information Technology, Florida landscape ecology, to provide novel fundamental insights into the dynamics and State University, Tallahassee, FL functioning of ecosystems from local to global scales, and to increase our ability to 32306-1100, USA predict the consequences of land-use changes on biodiversity and the provision of 3Department of Zoology, ecosystem services to human societies. University of Florida, 111 Bartram Hall, Gainesville, FL Keywords 32611-8525, -

Spatial Ecology and Modeling of Fish Populations - FAS 6416
Spatial Ecology and Modeling of Fish Populations - FAS 6416 1 Overview Spatial approaches to fisheries management are increasingly of interest the conservation and management of fish populations; and spatial research on fish populations and fish ecology is becoming a cornerstone of fisheries research. This course explores spatial ecology concepts, population dynamics models, examples of field data, and computer simulation tools to understand the ecological origin of spatial fish populations and their importance for fisheries management and conservation. The course is suitable for students who are interested in an interdisciplinary research field that draws as much from ecology as from fisheries management. 2 credits Spring Semester Face-to-face Location TBD with course website: http://elearning.ufl.edu/ The course is intended to provide students with different types of models and concepts that explain the spatial distribution of fish at different spatial and temporal scales. The course is new and evolving, and contains structured elements as well as exploratory exercises and discussions that are aimed at providing students with new perspectives on the dynamics of fish populations. Course Prerequisites: Graduate student status in Fisheries and Aquatic Sciences (or related, as determined by instructor). Interested graduate students in Wildlife Ecology are welcome. Instructor: Dr. Juliane Struve, [email protected]. SFRC Fisheries and Aquatic Sciences Rm. 34, Building 544. Telephone: 352-273-3632. Cell Phone: 352-213-5108 Please use the Canvas message/Inbox feature for fastest response. Office hours: Tuesdays, 3-5pm or online by appointment Textbook(s) and/or readings: The course does not have a designated text book. Reading materials will be assigned for every session and typical consist of 1-2 classic and recent papers. -

Landscapes of Fear: Spatial Patterns of Risk
TREE 2486 No. of Pages 14 Review Landscapes of Fear: Spatial Patterns of Risk Perception and Response 1, ,@ 2,3,5 1,5 4,5 Kaitlyn M. Gaynor , * Joel S. Brown, Arthur D. Middleton, Mary E. Power, and 1 Justin S. Brashares Animals experience varying levels of predation risk as they navigate heteroge- Highlights neous landscapes, and behavioral responses to perceived risk can structure Recent technological advances have provided unprecedented insights into ecosystems. The concept of the landscape of fear has recently become central the movement and behavior of animals to describing this spatial variation in risk, perception, and response. We present on heterogeneous landscapes. Some studies have indicated that spatial var- a framework linking the landscape of fear, defined as spatial variation in prey iation in predation risk plays a major perception of risk, to the underlying physical landscape and predation risk, and role in prey decision making, which can to resulting patterns of prey distribution and antipredator behavior. By disam- ultimately structure ecosystems. biguating the mechanisms through which prey perceive risk and incorporate The concept of the ‘landscape of fear’ fear into decision making, we can better quantify the nonlinear relationship was introduced in 2001 and has been between risk and response and evaluate the relative importance of the land- widely adopted to describe spatial var- iation predation risk, risk perception, scape of fear across taxa and ecosystems. and response. However, increasingly divergent interpretations of its meaning Introduction and application now cloud under- The risk of predation plays a powerful role in shaping behavior of fearful prey, with conse- standing and synthesis, and at least 15 distinct processes and states have quences for individual physiology, population dynamics, and community interactions [1,2]. -

Spatial Ecology of Territorial Populations
Spatial ecology of territorial populations Benjamin G. Weinera, Anna Posfaib, and Ned S. Wingreenc,1 aDepartment of Physics, Princeton University, Princeton, NJ 08544; bSimons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724; and cLewis–Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544 Edited by Nigel Goldenfeld, University of Illinois at Urbana–Champaign, Urbana, IL, and approved July 30, 2019 (received for review July 9, 2019) Many ecosystems, from vegetation to biofilms, are composed of and penalize niche overlap (25), but did not otherwise struc- territorial populations that compete for both nutrients and phys- ture the spatial interactions. All these models allow coexistence ical space. What are the implications of such spatial organization when the combination of spatial segregation and local interac- for biodiversity? To address this question, we developed and ana- tions weakens interspecific competition relative to intraspecifc lyzed a model of territorial resource competition. In the model, competition. However, it remains unclear how such interactions all species obey trade-offs inspired by biophysical constraints on relate to concrete biophysical processes. metabolism; the species occupy nonoverlapping territories, while Here, we study biodiversity in a model where species interact nutrients diffuse in space. We find that the nutrient diffusion through spatial resource competition. We specifically consider time is an important control parameter for both biodiversity and surface-associated populations which exclude each other as they the timescale of population dynamics. Interestingly, fast nutri- compete for territory. This is an appropriate description for ent diffusion allows the populations of some species to fluctuate biofilms, vegetation, and marine ecosystems like mussels (28) or to zero, leading to extinctions. -

Introduction to the Special Issue on Spatial Ecology
International Journal of Geo-Information Editorial Space-Ruled Ecological Processes: Introduction to the Special Issue on Spatial Ecology Duccio Rocchini 1,2,3 1 University of Trento, Center Agriculture Food Environment, Via E. Mach 1, 38010 S. Michele all’Adige (TN), Italy; [email protected] 2 University of Trento, Centre for Integrative Biology, Via Sommarive, 14, 38123 Povo (TN), Italy 3 Fondazione Edmund Mach, Department of Biodiversity and Molecular Ecology, Research and Innovation Centre, Via E. Mach 1, 38010 S. Michele all’Adige (TN), Italy Received: 12 December 2017; Accepted: 23 December 2017; Published: 2 January 2018 This special issue explores most of the scientific issues related to spatial ecology and its integration with geographical information at different spatial and temporal scales. Papers are mainly related to challenging aspects of species variability over space and landscape dynamics, providing a benchmark for future exploration on this theme. The need for a spatial view in Ecology is a fact. Dealing with ecological changes over space and time represents a long-lasting theme, now faced by means of innovative techniques and modelling approaches (e.g., Chaudhary et al. [1], Palmer [2], Rocchini [3]). This special issue explores some of them, with challenging ideas facing different components of spatial patterns related to ecological processes, such as: (i) species variability over space [4–11]; and (ii) landscape dynamics [12,13]. Concerning biodiversity variability over space, key spatial datasets are needed to fully investigate biodiversity change in space and time, as shown by Geri et al. [7], who carried out innovative procedures to recover and properly map historical floristic and vegetation data for biodiversity monitoring. -

Pattern and Process Second Edition Monica G. Turner Robert H. Gardner
Monica G. Turner Robert H. Gardner Landscape Ecology in Theory and Practice Pattern and Process Second Edition L ANDSCAPE E COLOGY IN T HEORY AND P RACTICE M ONICA G . T URNER R OBERT H . G ARDNER LANDSCAPE ECOLOGY IN THEORY AND PRACTICE Pattern and Process Second Edition Monica G. Turner University of Wisconsin-Madison Department of Zoology Madison , WI , USA Robert H. Gardner University of Maryland Center for Environmental Science Frostburg, MD , USA ISBN 978-1-4939-2793-7 ISBN 978-1-4939-2794-4 (eBook) DOI 10.1007/978-1-4939-2794-4 Library of Congress Control Number: 2015945952 Springer New York Heidelberg Dordrecht London © Springer-Verlag New York 2015 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifi cally the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfi lms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specifi c statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The publisher, the authors and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. Neither the publisher nor the authors or the editors give a warranty, express or implied, with respect to the material contained herein or for any errors or omissions that may have been made. -
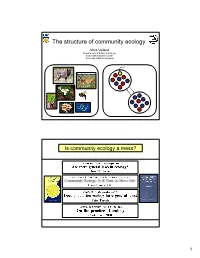
The Structure of Community Ecology
The structure of community ecology Mark Vellend Departments of Botany & Zoology Biodiversity Research Centre University of British Columbia Is community ecology a mess? 1 …with a key difference in how process is treated Population Community Genetics Ecology Patterns we Diversity and Diversity and want to composition of composition of understand alleles/genotypes species Competition Processes to Mutation Predation Niches Neutrality explain Drift Island biogeography Resource-use trade-offs patterns Colonization-competition trade-offs Migration Disturbance Productivity Evolutionary history Selection Etc. But is the difference real or necessary? Population Community Genetics Ecology Patterns we Diversity and Diversity and want to composition of composition of understand alleles/genotypes species Processes to Mutation Speciation explain Drift Drift patterns Migration Dispersal Selection Selection 2 Question on Ecology 101 final exam: What processes influence species composition and diversity? Answer: Question on Evolution 101 final exam: What processes influence genetic variation? Answer: Mutation Drift Migration Selection 3 Processes that influence community composition and diversity Speciation: The creation of new species. Drift: Random changes in the relative frequencies of species. Dispersal: Movement of organisms/propagules among communities. Selection: Deterministic differential survival or reproduction of species. THE LAST 40 The MacArthur school (Marlboro Circle) YEARS OF COMMUNITY Island Biogeography Simple, general, deterministic -

Spatial Ecology of Territorial Populations
bioRxiv preprint doi: https://doi.org/10.1101/694257; this version posted July 5, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Spatial Ecology of Territorial Populations Benjamin G. Weiner Department of Physics, Princeton University, Princeton, NJ 08544 Anna Posfai Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724 Ned S. Wingreen∗ Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544 (Dated: July 5, 2019) Many ecosystems, from vegetation to biofilms, are composed of territorial populations that com- pete for both nutrients and physical space. What are the implications of such spatial organization for biodiversity? To address this question, we developed and analyzed a model of territorial re- source competition. In the model, all species obey trade-offs inspired by biophysical constraints on metabolism; the species occupy non-overlapping territories while nutrients diffuse in space. We find that the nutrient diffusion time is an important control parameter for both biodiversity and the timescale of population dynamics. Interestingly, fast nutrient diffusion allows the populations of some species to fluctuate to zero, leading to extinctions. Moreover, territorial competition spon- taneously gives rise to both multistability and the Allee effect (in which a minimum population is required for survival), so that small perturbations can have major ecological effects. While the as- sumption of trade-offs allows for the coexistence of more species than the number of nutrients – thus violating the principle of competitive exclusion – overall biodiversity is curbed by the domination of “oligotroph” species. -

Wu-2012-LE-Encyclopedia TE.Pdf
L in real landscapes, and because it facilitates theoretical and methodological developments by recognizing the LANDSCAPE ECOLOGY importance of micro-, meso-, macro-, and cross-scale approaches. JIANGUO WU The term landscape ecology was coined in 1939 by Arizona State University the German geographer Carl Troll, who was inspired by the spatial patterning of landscapes revealed in aer- ial photographs and the ecosystem concept developed Spatial heterogeneity is ubiquitous in all ecological sys- in 1935 by the British ecologist Arthur Tansley. Troll tems, underlining the signifi cance of the pattern–process originally defi ned landscape ecology as the study of the relationship and the scale of observation and analysis. relationship between biological communities and their Landscape ecology focuses on the relationship between environment in a landscape mosaic. Today, landscape spatial pattern and ecological processes on multiple scales. ecology is widely recognized as the science of studying On the one hand, it represents a spatially explicit per- and improving the relationship between spatial pattern spective on ecological phenomena. On the other hand, and ecological processes on a multitude of scales and or- it is a highly interdisciplinary fi eld that integrates bio- ganizational levels. Heterogeneity, scale, pattern–process physical and socioeconomic perspectives to understand relationships, disturbance, hierarchy, and sustainability and improve the ecology and sustainability of landscapes. are among the key concepts in contemporary landscape Landscape ecology is still rapidly evolving, with a diver- ecology. Landscape ecological studies typically involve sity of emerging ideas and a plurality of methods and the use of geospatial data from various sources (e.g., applications. fi eld survey, aerial photography, and remote sensing) and spatial analysis of different kinds (e.g., pattern indi- DEFINING LANDSCAPE ECOLOGY ces and spatial statistics). -

Exploring Timescales of Predictability in Species Distributions
44 1–13 ECOGRAPHY Research Exploring timescales of predictability in species distributions Stephanie Brodie, Briana Abrahms, Steven J. Bograd, Gemma Carroll, Elliott L. Hazen, Barbara A. Muhling, Mercedes Pozo Buil, James A. Smith, Heather Welch and Michael G. Jacox S. Brodie (https://orcid.org/0000-0003-0869-9939) ✉ ([email protected]), S. J. Bograd, E. L. Hazen, B. A. Muhling, M. Pozo Buil, J. A. Smith, H. Welch and M. G. Jacox, Inst. of Marine Science, Univ. of California Santa Cruz, Santa Cruz, CA, USA. SBr, SBo, ELH, MPB, HW and MGJ also at: Environmental Research Division, NOAA Southwest Fisheries Science Center, Monterey, CA, USA. BAM and JAS also at: Fisheries Research Division, NOAA Southwest Fisheries Science Center, San Diego, CA, USA. – B. Abrahms, Center for Ecosystem Sentinels, Dept of Biology, Univ. of Washington, Seattle, WA, USA. – G. Carroll, School of Aquatic and Fisheries Sciences, Univ. of Washington, Seattle, WA, USA, and: Resource Ecology and Ecosystem Modelling Group, NOAA Alaska Fisheries Science Center, Seattle, WA, USA. Ecography Accurate forecasts of how animals respond to climate-driven environmental change 44: 1–13, 2021 are needed to prepare for future redistributions, however, it is unclear which temporal doi: 10.1111/ecog.05504 scales of environmental variability give rise to predictability of species distributions. We examined the temporal scales of environmental variability that best predicted Subject Editor: spatial abundance of a marine predator, swordfish Xiphias gladius, in the California Christine N. Meynard Current. To understand which temporal scales of environmental variability provide Editor-in-Chief: Miguel Araújo biological predictability, we decomposed physical variables into three components: a Accepted 16 February 2021 monthly climatology (long-term average), a low frequency component representing interannual variability, and a high frequency (sub-annual) component that captures ephemeral features. -

SWFSC Archive
Spatial Processes and Management of Marine Populations 667 Alaska Sea Grant College Program • AK-SG-01-02, 2001 Managing with Reserves: Modeling Uncertainty in Larval Dispersal for a Sea Urchin Fishery Lance E. Morgan National Marine Fisheries Service, Southwest Fisheries Science Center, Tiburon, California Louis W. Botsford University of California, Department of Wildlife, Fish and Conservation Biology, Davis, California Abstract The commercial fishery for the northern California red sea urchin, Strongylocentrotus franciscanus, began in 1985, peaked in 1989, and has declined steadily since that time. An alternative to traditional manage- ment is spatial management using no-take reserves that may improve the management of this and other fisheries. We formulated a size-structured metapopulation model with 24 patches and a dispersal matrix to address the key issue of larval linkages between reserve and fished patches. We explored four conceptual models of metapopulation connectivity includ- ing source-sink, limited distance, larval pool, and headlands dispersal models. These models indicated that the mode of dispersal led to poten- tially large differences in reserve performance. Larval pool models require little knowledge of spatial patterns in order to design an effective reserve network, while source-sink dynamics require a detailed understanding of larval dispersal if reserves are to be an effective means of fisheries man- agement. Reserve networks can be optimized with knowledge of larval dispersal patterns; however, they are rarely known with certainty. A deci- sion analysis used to evaluate alternative dispersal modes suggested a reserve system comprising approximately 25% of the total area can main- tain catch in the red sea urchin fishery, buffering uncertainty in the pat- tern of larval replenishment and fishing mortality rate. -

Manuscript Currently in Review at Ecological Applications (Updated June 2020)
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.09.142935; this version posted June 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Manuscript currently in review at Ecological Applications (Updated June 2020) 1 Living in the concrete jungle: carnivore spatial ecology in urban parks 2 3 4 Siria Gámez1, Nyeema C. Harris1 5 6 1Applied Wildlife Ecology Lab, Ecology and Evolutionary Biology, University of Michigan 7 1101 N. University Ave, Ann Arbor, Michigan 48106 8 9 Corresponding author: Siria Gámez ([email protected]) 10 11 ABSTRACT 12 13 People and wildlife are living in an increasingly urban world, replete with unprecedented human 14 densities, sprawling built environments, and altered landscapes. Such anthropogenic pressures 15 can affect processes at multiple ecological scales from individuals to ecosystems, yet few studies 16 integrate two or more levels of ecological organization. We tested two competing hypotheses, 17 humans as shields versus humans as competitors, to characterize how humans directly affect 18 carnivore spatial ecology across three scales. From 2017-2020, we conducted the first camera 19 survey of city parks in Detroit, Michigan, and obtained spatial occurrence data of the local native 20 carnivore community which included coyotes (Canis latrans), red foxes (Vulpes vulpes), gray 21 foxes (Urocyon cinereoargenteus), raccoons (Procyon lotor), and striped skunks (Mephitis 22 mephitis). We constructed single-species occupancy models to discriminate parks into areas of bioRxiv preprint doi: https://doi.org/10.1101/2020.06.09.142935; this version posted June 12, 2020.