Projections and Reformations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 3 Image Formation
This is page 44 Printer: Opaque this Chapter 3 Image Formation And since geometry is the right foundation of all painting, I have de- cided to teach its rudiments and principles to all youngsters eager for art... – Albrecht Durer¨ , The Art of Measurement, 1525 This chapter introduces simple mathematical models of the image formation pro- cess. In a broad figurative sense, vision is the inverse problem of image formation: the latter studies how objects give rise to images, while the former attempts to use images to recover a description of objects in space. Therefore, designing vision algorithms requires first developing a suitable model of image formation. Suit- able, in this context, does not necessarily mean physically accurate: the level of abstraction and complexity in modeling image formation must trade off physical constraints and mathematical simplicity in order to result in a manageable model (i.e. one that can be inverted with reasonable effort). Physical models of image formation easily exceed the level of complexity necessary and appropriate for this book, and determining the right model for the problem at hand is a form of engineering art. It comes as no surprise, then, that the study of image formation has for cen- turies been in the domain of artistic reproduction and composition, more so than of mathematics and engineering. Rudimentary understanding of the geometry of image formation, which includes various models for projecting the three- dimensional world onto a plane (e.g., a canvas), is implicit in various forms of visual arts. The roots of formulating the geometry of image formation can be traced back to the work of Euclid in the fourth century B.C. -

A Geometric Correction Method Based on Pixel Spatial Transformation
2020 International Conference on Computer Intelligent Systems and Network Remote Control (CISNRC 2020) ISBN: 978-1-60595-683-1 A Geometric Correction Method Based on Pixel Spatial Transformation Xiaoye Zhang, Shaobin Li, Lei Chen ABSTRACT To achieve the non-planar projection geometric correction, this paper proposes a geometric correction method based on pixel spatial transformation for non-planar projection. The pixel spatial transformation relationship between the distorted image on the non-planar projection plane and the original projection image is derived, the pre-distorted image corresponding to the projection surface is obtained, and the optimal view position is determined to complete the geometric correction. The experimental results show that this method can achieve geometric correction of projective plane as cylinder plane and spherical column plane, and can present the effect of visual distortion free at the optimal view point. KEYWORDS Geometric Correction, Image Pre-distortion, Pixel Spatial Transformation. INTRODUCTION Projection display technology can significantly increase visual range, and projection scenes can be flexibly arranged according to actual needs. Therefore, projection display technology has been applied to all aspects of production, life and learning. With the rapid development of digital media technology, modern projection requirements are also gradually complicated, such as virtual reality presentation with a high sense of immersion and projection performance in various forms, which cannot be met by traditional plane projection, so non-planar projection technology arises at the right moment. Compared with traditional planar projection display technology, non-planar projection mainly includes color compensation of projected surface[1], geometric splicing and brightness fusion of multiple projectors[2], and geometric correction[3]. -

CS 4204 Computer Graphics 3D Views and Projection
CS 4204 Computer Graphics 3D views and projection Adapted from notes by Yong Cao 1 Overview of 3D rendering Modeling: * Topic we’ve already discussed • *Define object in local coordinates • *Place object in world coordinates (modeling transformation) Viewing: • Define camera parameters • Find object location in camera coordinates (viewing transformation) Projection: project object to the viewplane Clipping: clip object to the view volume *Viewport transformation *Rasterization: rasterize object Simple teapot demo 3D rendering pipeline Vertices as input Series of operations/transformations to obtain 2D vertices in screen coordinates These can then be rasterized 3D rendering pipeline We’ve already discussed: • Viewport transformation • 3D modeling transformations We’ll talk about remaining topics in reverse order: • 3D clipping (simple extension of 2D clipping) • 3D projection • 3D viewing Clipping: 3D Cohen-Sutherland Use 6-bit outcodes When needed, clip line segment against planes Viewing and Projection Camera Analogy: 1. Set up your tripod and point the camera at the scene (viewing transformation). 2. Arrange the scene to be photographed into the desired composition (modeling transformation). 3. Choose a camera lens or adjust the zoom (projection transformation). 4. Determine how large you want the final photograph to be - for example, you might want it enlarged (viewport transformation). Projection transformations Introduction to Projection Transformations Mapping: f : Rn Rm Projection: n > m Planar Projection: Projection on a plane. -

Documenting Carved Stones by 3D Modelling
Documenting carved stones by 3D modelling – Example of Mongolian deer stones Fabrice Monna, Yury Esin, Jérôme Magail, Ludovic Granjon, Nicolas Navarro, Josef Wilczek, Laure Saligny, Sébastien Couette, Anthony Dumontet, Carmela Chateau Smith To cite this version: Fabrice Monna, Yury Esin, Jérôme Magail, Ludovic Granjon, Nicolas Navarro, et al.. Documenting carved stones by 3D modelling – Example of Mongolian deer stones. Journal of Cultural Heritage, El- sevier, 2018, 34 (November–December), pp.116-128. 10.1016/j.culher.2018.04.021. halshs-01916706 HAL Id: halshs-01916706 https://halshs.archives-ouvertes.fr/halshs-01916706 Submitted on 22 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/328759923 Documenting carved stones by 3D modelling-Example of Mongolian deer stones Article in Journal of Cultural Heritage · May 2018 DOI: 10.1016/j.culher.2018.04.021 CITATIONS READS 3 303 10 authors, including: Fabrice Monna Yury Esin University -

Lecture 16: Planar Homographies Robert Collins CSE486, Penn State Motivation: Points on Planar Surface
Robert Collins CSE486, Penn State Lecture 16: Planar Homographies Robert Collins CSE486, Penn State Motivation: Points on Planar Surface y x Robert Collins CSE486, Penn State Review : Forward Projection World Camera Film Pixel Coords Coords Coords Coords U X x u M M V ext Y proj y Maff v W Z U X Mint u V Y v W Z U M u V m11 m12 m13 m14 v W m21 m22 m23 m24 m31 m31 m33 m34 Robert Collins CSE486, PennWorld State to Camera Transformation PC PW W Y X U R Z C V Rotate to Translate by - C align axes (align origins) PC = R ( PW - C ) = R PW + T Robert Collins CSE486, Penn State Perspective Matrix Equation X (Camera Coordinates) x = f Z Y X y = f x' f 0 0 0 Z Y y' = 0 f 0 0 Z z' 0 0 1 0 1 p = M int ⋅ PC Robert Collins CSE486, Penn State Film to Pixel Coords 2D affine transformation from film coords (x,y) to pixel coordinates (u,v): X u’ a11 a12xa'13 f 0 0 0 Y v’ a21 a22 ya'23 = 0 f 0 0 w’ Z 0 0z1' 0 0 1 0 1 Maff Mproj u = Mint PC = Maff Mproj PC Robert Collins CSE486, Penn StateProjection of Points on Planar Surface Perspective projection y Film coordinates x Point on plane Rotation + Translation Robert Collins CSE486, Penn State Projection of Planar Points Robert Collins CSE486, Penn StateProjection of Planar Points (cont) Homography H (planar projective transformation) Robert Collins CSE486, Penn StateProjection of Planar Points (cont) Homography H (planar projective transformation) Punchline: For planar surfaces, 3D to 2D perspective projection reduces to a 2D to 2D transformation. -

Projections in Context
Projections in Context Kaye Mason and Sheelagh Carpendale and Brian Wyvill Department of Computer Science, University of Calgary Calgary, Alberta, Canada g g ffkatherim—sheelagh—blob @cpsc.ucalgary.ca Abstract sentation is the technical difficulty in the more mathematical as- pects of projection. This is exactly where computers can be of great This examination considers projections from three space into two assistance to the authors of representations. It is difficult enough to space, and in particular their application in the visual arts and in get all of the angles right in a planar geometric projection. Get- computer graphics, for the creation of image representations of the ting the curves right in a non-planar projection requires a great deal real world. A consideration of the history of projections both in the of skill. An algorithm, however, could provide a great deal of as- visual arts, and in computer graphics gives the background for a sistance. A few computer scientists have realised this, and begun discussion of possible extensions to allow for more author control, work on developing a broader conceptual framework for the imple- and a broader range of stylistic support. Consideration is given mentation of projections, and projection-aiding tools in computer to supporting user access to these extensions and to the potential graphics. utility. This examination considers this problem of projecting a three dimensional phenomenon onto a two dimensional media, be it a Keywords: Computer Graphics, Perspective, Projective Geome- canvas, a tapestry or a computer screen. It begins by examining try, NPR the nature of the problem (Section 2) and then look at solutions that have been developed both by artists (Section 3) and by com- puter scientists (Section 4). -

Projector Pixel Redirection Using Phase-Only Spatial Light Modulator
Projector Pixel Redirection Using Phase-Only Spatial Light Modulator Haruka Terai, Daisuke Iwai, and Kosuke Sato Abstract—In projection mapping from a projector to a non-planar surface, the pixel density on the surface becomes uneven. This causes the critical problem of local spatial resolution degradation. We confirmed that the pixel density uniformity on the surface was improved by redirecting projected rays using a phase-only spatial light modulator. Index Terms—Phase-only spatial light modulator (PSLM), pixel redirection, pixel density uniformization 1 INTRODUCTION Projection mapping is a technology for superimposing an image onto a three-dimensional object with an uneven surface using a projector. It changes the appearance of objects such as color and texture, and is used in various fields, such as medical [4] or design support [5]. Projectors are generally designed so that the projected pixel density is uniform when an image is projected on a flat screen. When an image is projected on a non-planar surface, the pixel density becomes uneven, which leads to significant degradation of spatial resolution locally. Previous works applied multiple projectors to compensate for the local resolution degradation. Specifically, projectors are placed so that they illuminate a target object from different directions. For each point on the surface, the previous techniques selected one of the projectors that can project the finest image on the point. In run-time, each point is illuminated by the selected projector [1–3]. Although these techniques Fig. 1. Phase modulation principle by PSLM-LCOS. successfully improved the local spatial degradation, their system con- figurations are costly and require precise alignment of projected images at sub-pixel accuracy. -

How Useful Is Projective Geometry? Patrick Gros, Richard Hartley, Roger Mohr, Long Quan
How Useful is Projective Geometry? Patrick Gros, Richard Hartley, Roger Mohr, Long Quan To cite this version: Patrick Gros, Richard Hartley, Roger Mohr, Long Quan. How Useful is Projective Geometry?. Com- puter Vision and Image Understanding, Elsevier, 1997, 65 (3), pp.442–446. 10.1006/cviu.1996.0496. inria-00548360 HAL Id: inria-00548360 https://hal.inria.fr/inria-00548360 Submitted on 22 Dec 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. How useful is pro jective geometry 1 1 1 2 P Gros R Mohr L Quan R Hartley 1 LIFIA INRIA RhoneAlp es avenue F Viallet Grenoble Cedex France 2 GE CRD Schenectady NY USA In this resp onse we will weigh two dierent approaches to shap e recognition On the one hand we have the use of restricted camera mo dels as advo cated in the pap er of Pizlo et al to give a closer approximation to real calibrated cameras The alternative approach is to use a full pro jective camera mo del and take advantage of the machinery of pro jective geometry Ab out the p ersp ective pro jection Before discussing the usefulness -

Chapter 5 Reconstruction from Two Calibrated Views
This is page 109 Printer: Opaque this Chapter 5 Reconstruction from Two Calibrated Views We see because we move; we move because we see. – James J. Gibson, The Perception of the Visual World In this chapter we begin unveiling the basic geometry that relates images of points to their 3-D position. We start with the simplest case of two calibrated cam- eras, and describe an algorithm, first proposed by the British psychologist H.C. Longuet-Higgins in 1981, to reconstruct the relative pose (i.e. position and ori- entation) of the cameras as well as the locations of the points in space from their projection onto the two images. It has been long known in photogrammetry that the coordinates of the projec- tion of a point and the two camera optical centers form a triangle (Figure 5.1), a fact that can be written as an algebraic constraint involving the camera poses and image coordinates but not the 3-D position of the points. Given enough points, therefore, this constraint can be used to solve for the camera poses. Once those are known, the 3-D position of the points can be obtained easily by triangula- tion. The interesting feature of the constraint is that although it is nonlinear in the unknown camera poses, it can be solved by two linear steps in closed form. Therefore, in the absence of any noise or uncertainty, given two images taken from calibrated cameras, one can in principle recover camera pose and position of the points in space with a few steps of simple linear algebra. -
Cs-174-Lecture-Projections.Pdf
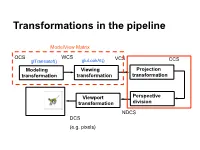
Transformations in the pipeline ModelView Matrix OCS WCS VCS glTranslatef() gluLookAt() CCS … Modeling Viewing Projection transformation transformation transformation Viewport Perspective transformation division NDCS DCS (e.g. pixels) Background (reminder) Line (in 2D) • Explicit • Implicit • Parametric Background (reminder) Plane equations Implicit Parametric Explicit Reminder: Homogeneous Coordinates Projection transformations Introduction to Projection Transformations [Hill: 371-378, 398-404. Foley & van Dam: p. 229-242 ] Mapping: f : Rn Rm Projection: n > m Planar Projection: Projection on a plane. R3R2 or R4R3 homogenous coordinates. Basic projections Parallel Perspective Taxonomy Examples A basic orthographic projection x’ = x y’ = y z’ = N Y Matrix Form N Z I A basic perspective projection Similar triangles x’/d = x/(-z) -> x’ = x d/(-z) y’/d = y/(-z) => y’ = y d/(-z) z’ = -d In matrix form Matrix form of x’ = x d/(-z) y’ = y d/(-z) z’ = -d Moving from 4D to 3D Projections in OpenGL Camera coordinate system Image plane = near plane Camera at (0,0,0) Looking at –z Image plane at z = -N Perspective projection of a point In eye coordinates P =[Px,Py,Pz,1]T x/Px = N/(-Pz) => x = NPx/(-Pz) y/Px = N/(-Pz) => y = NPy/(-Pz) Observations • Perspective foreshortening • Denominator becomes undefined for z = 0 • If P is behind the eye Pz changes sign • Near plane just scales the picture • Straight line -> straight line Perspective projection of a line Perspective Division, drop fourth coordinate Is it a line? Cont’d next slide Is it a line? -

Three Dimensional Occlusion Mapping Using a Lidar Sensor
Freie Universität Berlin Fachbereich Mathematik und Informatik Masterarbeit Three dimensional occlusion mapping using a LiDAR sensor von: David Damm Matrikelnummer: 4547660 Email: [email protected] Betreuer: Prof. Dr. Daniel Göhring und Fritz Ulbrich Berlin, 30.08.2018 Abstract Decision-making is an important task in autonomous driving. Especially in dy- namic environments, it is hard to be aware of every unpredictable event that would affect the decision. To face this issue, an autonomous car is equipped with different types of sensors. The LiDAR laser sensors, for example, can create a three dimensional 360 degree representation of the environment with precise depth information. But like humans, the laser sensors have their lim- itations. The field of view could be partly blocked by a truck and with that, the area behind the truck would be completely out of view. These occluded areas in the environment increase the uncertainty of the decision and ignoring this uncertainty would increase the risk of an accident. This thesis presents an approach to estimate such areas from the data of a LiDAR sensor. Therefore, different methods will be discussed and finally the preferred method evalu- ated. Statement of Academic Integrity Hereby, I declare that I have composed the presented paper independently on my own and without any other resources than the ones indicated. All thoughts taken directly or indirectly from external sources are properly denoted as such. This paper has neither been previously submitted to another authority nor has it been published yet. August 30, 2018 David Damm Contents Contents 1 Introduction 1 1.1 Motivation . -

IMAGE-BASED MODELING TECHNIQUES for ARTISTIC RENDERING Bynum Murray Iii Clemson University, [email protected]
Clemson University TigerPrints All Theses Theses 5-2010 IMAGE-BASED MODELING TECHNIQUES FOR ARTISTIC RENDERING Bynum Murray iii Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_theses Part of the Fine Arts Commons Recommended Citation Murray iii, Bynum, "IMAGE-BASED MODELING TECHNIQUES FOR ARTISTIC RENDERING" (2010). All Theses. 777. https://tigerprints.clemson.edu/all_theses/777 This Thesis is brought to you for free and open access by the Theses at TigerPrints. It has been accepted for inclusion in All Theses by an authorized administrator of TigerPrints. For more information, please contact [email protected]. IMAGE-BASED MODELING TECHNIQUES FOR ARTISTIC RENDERING A Thesis Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Master of Arts Digital Production Arts by Bynum Edward Murray III May 2010 Accepted by: Timothy Davis, Ph.D. Committee Chair David Donar, M.F.A. Tony Penna, M.F.A. ABSTRACT This thesis presents various techniques for recreating and enhancing two- dimensional paintings and images in three-dimensional ways. The techniques include camera projection modeling, digital relief sculpture, and digital impasto. We also explore current problems of replicating and enhancing natural media and describe various solutions, along with their relative strengths and weaknesses. The importance of artistic skill in the implementation of these techniques is covered, along with implementation within the current industry applications Autodesk Maya, Adobe Photoshop, Corel Painter, and Pixologic Zbrush. The result is a set of methods for the digital artist to create effects that would not otherwise be possible.