The Key to Translational Research J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Intraocular Pressure Rise After Phacoemulsification Prophylactic
British Journal of Ophthalmology 1995; 79: 809-813 809 Intraocular pressure rise after phacoemulsification Br J Ophthalmol: first published as 10.1136/bjo.79.9.809 on 1 September 1995. Downloaded from with posterior chamber lens implantation: effect of prophylactic medication, wound closure, and surgeon's experience Thomas G Bomer, Wolf-Dietrich A Lagreze, Jens Funk Abstract pressure rises usually occur between 6 and 8 Aims-A prospective clinical trial was hours after surgery.6 carried out to evaluate the effect of Various antiglaucomatous agents have been prophylactic medication, the technique of used to prevent the intraocular pressure rise wound closure, and the surgeon's experi- after cataract extraction. Oral acetazolamide15 16 ence on the intraocular pressure rise after and topical timolol'6-19 lowered the pressure cataract extraction. rise in the early period after intracapsular and Methods-In 100 eyes, the intraocular extracapsular cataract extraction. Levobunolol pressure was measured before as well as proved to be superior to timolol 4-7 hours 2-4, 5-7, and 22-24 hours after phaco- after extracapsular cataract extraction.20 Apra- emulsification and posterior chamber lens clonidine lowered the intraocular pressure rise implantation. Each of 25 patients received after uncomplicated phacoemulsification21 and either 1% topical apraclonidine, 0.5%/o extracapsular cataract extraction22 23 when topical levobunolol, 500 mg oral acetazo- given 30 minutes to 1 hour before surgery, lamide, or placebo. Forty four eyes were whereas immediate postoperative treatment operated with sclerocorneal sutureless with apraclonidine was ineffective.22 24 Miotics tunnel and 56 eyes with corneoscleral are frequently used to promote miosis and incision and suture. -

Brimonidine Tartrate; Brinzolamide
Contains Nonbinding Recommendations Draft Guidance on Brimonidine Tartrate ; Brinzolamide This draft guidance, when finalized, will represent the current thinking of the Food and Drug Administration (FDA, or the Agency) on this topic. It does not establish any rights for any person and is not binding on FDA or the public. You can use an alternative approach if it satisfies the requirements of the applicable statutes and regulations. To discuss an alternative approach, contact the Office of Generic Drugs. Active Ingredient: Brimonidine tartrate; Brinzolamide Dosage Form; Route: Suspension/drops; ophthalmic Strength: 0.2%; 1% Recommended Studies: One study Type of study: Bioequivalence (BE) study with clinical endpoint Design: Randomized (1:1), double-masked, parallel, two-arm, in vivo Strength: 0.2%; 1% Subjects: Males and females with chronic open angle glaucoma or ocular hypertension in both eyes. Additional comments: Specific recommendations are provided below. ______________________________________________________________________________ Analytes to measure (in appropriate biological fluid): Not applicable Bioequivalence based on (95% CI): Clinical endpoint Additional comments regarding the BE study with clinical endpoint: 1. The Office of Generic Drugs (OGD) recommends conducting a BE study with a clinical endpoint in the treatment of open angle glaucoma and ocular hypertension comparing the test product to the reference listed drug (RLD), each applied as one drop in both eyes three times daily at approximately 8:00 a.m., 4:00 p.m., and 10:00 p.m. for 42 days (6 weeks). 2. Inclusion criteria (the sponsor may add additional criteria): a. Male or nonpregnant females aged at least 18 years with chronic open angle glaucoma or ocular hypertension in both eyes b. -

The In¯Uence of Medication on Erectile Function
International Journal of Impotence Research (1997) 9, 17±26 ß 1997 Stockton Press All rights reserved 0955-9930/97 $12.00 The in¯uence of medication on erectile function W Meinhardt1, RF Kropman2, P Vermeij3, AAB Lycklama aÁ Nijeholt4 and J Zwartendijk4 1Department of Urology, Netherlands Cancer Institute/Antoni van Leeuwenhoek Hospital, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; 2Department of Urology, Leyenburg Hospital, Leyweg 275, 2545 CH The Hague, The Netherlands; 3Pharmacy; and 4Department of Urology, Leiden University Hospital, P.O. Box 9600, 2300 RC Leiden, The Netherlands Keywords: impotence; side-effect; antipsychotic; antihypertensive; physiology; erectile function Introduction stopped their antihypertensive treatment over a ®ve year period, because of side-effects on sexual function.5 In the drug registration procedures sexual Several physiological mechanisms are involved in function is not a major issue. This means that erectile function. A negative in¯uence of prescrip- knowledge of the problem is mainly dependent on tion-drugs on these mechanisms will not always case reports and the lists from side effect registries.6±8 come to the attention of the clinician, whereas a Another way of looking at the problem is drug causing priapism will rarely escape the atten- combining available data on mechanisms of action tion. of drugs with the knowledge of the physiological When erectile function is in¯uenced in a negative mechanisms involved in erectile function. The way compensation may occur. For example, age- advantage of this approach is that remedies may related penile sensory disorders may be compen- evolve from it. sated for by extra stimulation.1 Diminished in¯ux of In this paper we will discuss the subject in the blood will lead to a slower onset of the erection, but following order: may be accepted. -

Properties and Units in Clinical Pharmacology and Toxicology
Pure Appl. Chem., Vol. 72, No. 3, pp. 479–552, 2000. © 2000 IUPAC INTERNATIONAL FEDERATION OF CLINICAL CHEMISTRY AND LABORATORY MEDICINE SCIENTIFIC DIVISION COMMITTEE ON NOMENCLATURE, PROPERTIES, AND UNITS (C-NPU)# and INTERNATIONAL UNION OF PURE AND APPLIED CHEMISTRY CHEMISTRY AND HUMAN HEALTH DIVISION CLINICAL CHEMISTRY SECTION COMMISSION ON NOMENCLATURE, PROPERTIES, AND UNITS (C-NPU)§ PROPERTIES AND UNITS IN THE CLINICAL LABORATORY SCIENCES PART XII. PROPERTIES AND UNITS IN CLINICAL PHARMACOLOGY AND TOXICOLOGY (Technical Report) (IFCC–IUPAC 1999) Prepared for publication by HENRIK OLESEN1, DAVID COWAN2, RAFAEL DE LA TORRE3 , IVAN BRUUNSHUUS1, MORTEN ROHDE1, and DESMOND KENNY4 1Office of Laboratory Informatics, Copenhagen University Hospital (Rigshospitalet), Copenhagen, Denmark; 2Drug Control Centre, London University, King’s College, London, UK; 3IMIM, Dr. Aiguader 80, Barcelona, Spain; 4Dept. of Clinical Biochemistry, Our Lady’s Hospital for Sick Children, Crumlin, Dublin 12, Ireland #§The combined Memberships of the Committee and the Commission (C-NPU) during the preparation of this report (1994–1996) were as follows: Chairman: H. Olesen (Denmark, 1989–1995); D. Kenny (Ireland, 1996); Members: X. Fuentes-Arderiu (Spain, 1991–1997); J. G. Hill (Canada, 1987–1997); D. Kenny (Ireland, 1994–1997); H. Olesen (Denmark, 1985–1995); P. L. Storring (UK, 1989–1995); P. Soares de Araujo (Brazil, 1994–1997); R. Dybkær (Denmark, 1996–1997); C. McDonald (USA, 1996–1997). Please forward comments to: H. Olesen, Office of Laboratory Informatics 76-6-1, Copenhagen University Hospital (Rigshospitalet), 9 Blegdamsvej, DK-2100 Copenhagen, Denmark. E-mail: [email protected] Republication or reproduction of this report or its storage and/or dissemination by electronic means is permitted without the need for formal IUPAC permission on condition that an acknowledgment, with full reference to the source, along with use of the copyright symbol ©, the name IUPAC, and the year of publication, are prominently visible. -

Table 1. Glaucoma Medications: Mechanisms, Dosing and Precautions Brand Generic Mechanism of Action Dosage/Avg
OPTOMETRIC STUDY CENTER Table 1. Glaucoma Medications: Mechanisms, Dosing and Precautions Brand Generic Mechanism of Action Dosage/Avg. % Product Sizes Side Effects Warnings Reduction CHOLINERGIC AGENTS Direct Pilocarpine (generic) Pilocarpine 1%, 2%, 4% Increases trabecular outflow BID-QID/15-25% 15ml Headache, blurred vision, myopia, retinal detachment, bronchiole constriction, Angle closure, shortness of breath, retinal narrowing of angle detachment Indirect Phospholine Iodide (Pfizer) Echothiophate iodide 0.125% Increases trabecular outflow QD-BID/15-25% 5ml Same as above plus cataractogenic iris cysts in children, pupillary block, Same as above, plus avoid prior to any increased paralysis with succinylcholine general anesthetic procedure ALPHA-2 AGONISTS Alphagan P (Allergan) Brimonidine tartrate 0.1%, 0.15% with Purite Decreases aqueous production, increases BID-TID/up to 26% 5ml, 10ml, 15ml Dry mouth, hypotension, bradycardia, follicular conjunctivitis, ocular irritation, Monitor for shortness of breath, dizziness, preservative uveoscleral outflow pruritus, dermatitis, conjunctival blanching, eyelid retraction, mydriasis, drug ocular redness and itching, fatigue allergy Brimonidine tartrate Brimonidine tartrate 0.15%, 0.2% Same as above Same as above 5ml, 10ml Same as above Same as above (generic) Iopidine (Novartis) Apraclonidine 0.5% Decreases aqueous production BID-TID/up to 25% 5ml, 10ml Same as above but higher drug allergy (40%) Same as above BETA-BLOCKERS Non-selective Betagan (Allergan) Levobunolol 0.25%, 0.5% Decreases -

NEW ZEALAND DATA SHEET 1. PRODUCT NAME IOPIDINE® (Apraclonidine Hydrochloride) Eye Drops 0.5%
NEW ZEALAND DATA SHEET 1. PRODUCT NAME IOPIDINE® (apraclonidine hydrochloride) Eye Drops 0.5%. 2. QUALITATIVE AND QUANTITATIVE COMPOSITION Each mL of Iopidine Eye Drops 0.5% contains apraclonidine hydrochloride 5.75 mg, equivalent to apraclonidine base 5 mg. Excipient with known effect Benzalkonium chloride 0.1 mg per 1 mL as a preservative. For the full list of excipients, see section 6.1. 2. PHARMACEUTICAL FORM Eye drops, solution, sterile, isotonic. 4. CLINICAL PARTICULARS 4.1. Therapeutic indications Iopidine Eye Drops 0.5% are indicated for short-term adjunctive therapy in patients on maximally tolerated medical therapy who require additional IOP reduction. Patients on maximally tolerated medical therapy who are treated with Iopidine Eye Drops 0.5% to delay surgery should have frequent follow up examinations and treatment should be discontinued if the intraocular pressure rises significantly. The addition of Iopidine Eye Drops 0.5% to patients already using two aqueous suppressing drugs (i.e. beta-blocker plus carbonic anhydrase inhibitor) as part of their maximally tolerated medical therapy may not provide additional benefit. This is because apraclonidine is an aqueous-suppressing drug and the addition of a third aqueous suppressant may not significantly reduce IOP. The IOP-lowering efficacy of Iopidine Eye Drops 0.5% diminishes over time in some patients. This loss of effect, or tachyphylaxis, appears to be an individual occurrence with a variable time of onset and should be closely monitored. The benefit for most patients is less than one month. 4.2. Dose and method of administration Dose One drop of Iopidine Eye Drops 0.5% should be instilled into the affected eye(s) three times per day. -

Hyperlinked Region IX SOP for MWLCEMS
2 Region IX 2 0 0 STANDARD OPERATING PROCEDURES/ 1 1 STANDING MEDICAL ORDERS 9 9 McHenry Western Lake County EMS System Healthcare delivery requires structure (people, equipment, education) and process (policies, protocols, procedures) that, when integrated, produce a system (programs, organizations, cultures) that leads to optimal outcomes (patient survival and safety, quality, satisfaction). An effective system of care comprises all of these elements—structure, process, system, and patient outcomes—in a framework of continuous quality improvement (AHA, 2015). These protocols have been developed and approved through a collaborative process involving the Advocate Lutheran General; Greater Elgin Area, McHenry Western Lake County, Northwest Community, Amita Saint Joseph Hospital, and Southern Fox Valley EMS Systems to reduce variation in practice and establish a Region-wide System of care. They shall be used: as the written practice guidelines/pathways of care approved by the EMS Medical Directors (EMS MDs) to be initiated by System EMS personnel for off-line medical control. as the standing medical orders to be used by Emergency Communications Registered Nurses (ECRNs) when providing on-line medical control (OLMC). in medium to large scale multiple patient incidents, given that the usual and customary forms of communication are contraindicated as specified in the Region IX disaster plan. System members are authorized to implement these orders to their scope of practice. OLMC communication shall be established without endangering the patient. Under no circumstances shall emergency prehospital care be delayed while attempting to establish contact with a hospital. In the event that communications cannot be established, EMS personnel shall continue to provide care to the degree authorized by their license, these protocols, drugs/equipment available, and their scope of practice granted by the EMS MD in that System. -

0Bcore Safety Profile
Core Safety Profile Active substance: Levobunolol Pharmaceutical form(s)/strength: Eye drops solution/ 0,1%; 0,25%; 0,5%; 0,5% UD P-RMS: CZ/H/PSUR/0006/001 Date of FAR: 26.05.2009 4.2 Posology and method of administration Adults (including the elderly) Country specific posology and method of administration to be included. Children /.../ is not recommended for use in children due to lack of safety and efficacy data. If required, /.../ may be used with other agents to lower intra-ocular pressure. The use of two topical beta-adrenergic blocking agents is not recommended (see section 4.4). Intraocular pressure should be measured approximately four weeks after starting treatment with /.../ as a return to normal ocular pressure can take a few weeks. As with any eye drops, to reduce possible systemic absorption, it is recommended that the lachrymal sac is compressed at the medial canthus (punctual occlusion) for one minute. This should be performed immediately following the instillation of each drop. Transfer from other beta-blocking treatment When another beta blocking agent is being used treatment must be discontinued after a full day of therapy. Start treatment with /.../ the next day with X drop of /.../ topically applied into the conjunctival sac in the affected eye(s). If /.../ is to replace a combination of anti-glaucoma products, only a single product should be removed at a time. Use in renal and hepatic impairment Levobunolol hydrochloride has not been studied in patients with hepatic or renal impairment. Therefore, caution should be used in treating such patients (see section 4.4). -

Sustained-Release Compositions Containing Cation Exchange Resins and Polycarboxylic Polymers
~" ' Nil II II II II Ml Ml INI MINI II J European Patent Office *»%r\ n » © Publication number: 0 429 732 B1 Office_„. europeen des brevets © EUROPEAN PATENT SPECIFICATION © Date of publication of patent specification: 16.03.94 © Int. CI.5: A61 K 9/06, A61 K 9/1 8, A61 K 47/32 © Application number: 89312590.6 @ Date of filing: 01.12.89 © Sustained-release compositions containing cation exchange resins and polycarboxylic polymers. @ Date of publication of application: (73) Proprietor: ALCON LABORATORIES, INC. 05.06.91 Bulletin 91/23 6201 South Freeway Fort Worth Texas 76107(US) © Publication of the grant of the patent: 16.03.94 Bulletin 94/11 @ Inventor: Janl, Rajni 4621 Briarhaven Road © Designated Contracting States: Fort Worth, Texas 76109(US) AT BE CH DE FR GB IT LI LU NL SE Inventor: Harris, Robert Gregg 3224 Westcliff Road W. © References cited: Fort Worth, Texas 76109(US) EP-A- 0 254 822 J.Pharm. SCI., vol. 60, no. 9, September 1971, © Representative: Jump, Timothy John Simon et pages 1343-1345 A. HEYD:"Polymer-drug in- al teraction: stability of aqueous gels contain- Venner Shipley & Co. ing neomycin sulfate" 20 Little Britain London EC1A 7DH (GB) 00 CM CO o> CM Note: Within nine months from the publication of the mention of the grant of the European patent, any person ® may give notice to the European Patent Office of opposition to the European patent granted. Notice of opposition CL shall be filed in a written reasoned statement. It shall not be deemed to have been filed until the opposition fee LU has been paid (Art. -
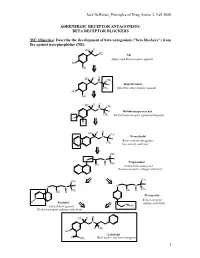
BETA RECEPTOR BLOCKERS MC Objective
Jack DeRuiter, Principles of Drug Action 2, Fall 2000 ADRENERGIC RECEPTOR ANTAGONISTS: BETA RECEPTOR BLOCKERS MC Objective: Describe the development of beta antagonists ("beta blockers") from the agonist norepinephrine (NE): HO H NH2 NE Alpha- and Beta-receptor agonist HO OH H H HO CH N 3 H Isoproterenol CH3 Selective beta-receptor agonist HO OH H H HO CH N 3 H Dichloroisoproterenol CH3 Partial beta-receptor agonist/antagonist Cl Cl H H HO CH N 3 Pronethalol H Beta-receptor antagonist, CH 3 low activity and toxic CH3 O N H Propranolol CH3 OH H Potent beta-antagonist No beta-receptor subtype selectivity CH3 CH3 O N H O N H CH CH OH H 3 OH H 3 Metoprolol N Beta-1-receptor H Pindolol O subtype selectivity Partial beta-agonist CH3 No beta-receptor subtype selectivity HO H H N H CH3 HO Labetolol O NH2 Dual alpha- and beta-antagonst 1 Jack DeRuiter, Principles of Drug Action 2, Fall 2000 MC Objective: Based on their structures, would the beta-blockers be expected to be relatively receptor selective? YES. They do not produce significant blockade of alpha- adrenergic receptors (alpha-1 or alpha-2), histamine receptors, muscarinic receptors or dopamine receptors. MC/PC Objective: Identify which beta blockers are classified as "non-selective": · The “non-selective" classification refers to those beta-blockers capable of blocking BOTH beta-1 and beta-2 receptors with equivalent efficacy. These drugs DO NOT have clinically significant affinity for other neurotransmitter receptors (alpha, dopamine, histamine, acetylcholine, etc.). · ALL of these beta-blockers (except satolol) consist of an aryloxypropanolamine side chain linked to an aromatic or “heteroaromatic” ring which is “ortho” substituted. -

Antiglaucoma Compositions Containing Combinations of Alpha-2 Agonists and Beta-Blockers
~" ' MM II II II MM II II II Ml II II I II J European Patent Office _ _ _ © Publication number: 0 365 662 B1 Office europeen* des.. brevets , © EUROPEAN PATENT SPECIFICATION © Date of publication of patent specification: 09.03.94 © Int. CI.5: A61 K 31/47 © Application number: 89905874.7 @ Date of filing: 26.04.89 © International application number: PCT/US89/01994 © International publication number: WO 89/10126 (02.11.89 89/26) (54) ANTIGLAUCOMA COMPOSITIONS CONTAINING COMBINATIONS OF ALPHA-2 AGONISTS AND BETA-BLOCKERS. ® Priority: 26.04.88 US 186504 1075-1078 @ Date of publication of application: J. of Cardiovascular Pharmacology, vol. 2, 02.05.90 Bulletin 90/18 suppl. 1, 1980, S. 21-28 © Publication of the grant of the patent: © Proprietor: ALCON LABORATORIES INC 09.03.94 Bulletin 94/10 6201 South Freeway Ft. Worth, TX 76134(US) © Designated Contracting States: AT BE CH DE FR GB IT LI LU NL SE @ Inventor: DE SANTIS, Louis, M. 2316 Wlnton Terrace West © References cited: Fort Worth, TX 761 09(US) US-A- 4 455 317 US-A- 4 515 800 © Representative: Jump, Timothy John Simon et AM A DRUG EVALUATION, 2nd ed., 1973; al "Agent used to treat Glaucoma", pp. 675-686. Venner Shipley & Co. 20 Little Britain GLAUCOMA, vol. 1, no. 1, February 1979; London EC1A 7DH (GB) 00 S.SUGAR, pp. 9-15. CM CO Ophthalmology, vol. 96, no. 1, 1989, p. 3-7 CO m Ophthalmology, vol. 98, no. 7, 1991, p. CO 00 Note: Within nine months from the publication of the mention of the grant of the European patent, any person may give notice to the European Patent Office of opposition to the European patent granted. -

REMEDY for GLAUCOMA COMPRISING Rho KINASE INHIBITOR and -BLOCKER
Europäisches Patentamt *EP001568382A1* (19) European Patent Office Office européen des brevets (11) EP 1 568 382 A1 (12) EUROPEAN PATENT APPLICATION published in accordance with Art. 158(3) EPC (43) Date of publication: (51) Int Cl.7: A61K 45/06, A61K 31/138, 31.08.2005 Bulletin 2005/35 A61K 31/343, A61K 31/353, A61K 31/437, A61K 31/4409, (21) Application number: 03772800.3 A61K 31/4412, A61K 31/496, (22) Date of filing: 17.11.2003 A61K 31/5377, A61K 31/551, A61P 27/06, A61P 43/00 (86) International application number: PCT/JP2003/014559 (87) International publication number: WO 2004/045644 (03.06.2004 Gazette 2004/23) (84) Designated Contracting States: • NAKAJIMA, Tadashi AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Ikoma-shi, Nara 630-0101 (JP) HU IE IT LI LU MC NL PT RO SE SI SK TR • MATSUGI, Takeshi Designated Extension States: Ikoma-shi, Nara 630-0101 (JP) AL LT LV MK • HARA, Hideaki Ikoma-shi, Nara 630-0101 (JP) (30) Priority: 18.11.2002 JP 2002333213 (74) Representative: Peaucelle, Chantal et al (71) Applicant: SANTEN PHARMACEUTICAL CO., Cabinet Armengaud Ainé LTD. 3, avenue Bugeaud Osaka 533-8651 (JP) 75116 Paris (FR) (72) Inventors: • HATANO, Masakazu Ikoma-shi, Nara 630-0101 (JP) (54) REMEDY FOR GLAUCOMA COMPRISING Rho KINASE INHIBITOR AND &bgr;-BLOCKER (57) A subject of the present invention is to find utility of a co mbination of a Rho kinase inhibitor having a novel action and a β-blocker as a therapeutic agent for glau- coma.