Virology Journal Biomed Central
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tamada-Text R3 HK-TT
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Okayama University Scientific Achievement Repository Tamada & Kondo - 1 Journal of General Plant Pathology Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors Tetsuo Tamada* and Hideki Kondo Institute of Plant Science and Resources (IPSR), Okayama University, Kurashiki 710-0046, Japan. Current address for T. Tamada: Kita 10, Nishi 1, 13-2-606, Sapporo, 001-0010, Japan. *Corresponding author: Tetsuo Tamada; E-mail: [email protected] Total text pages 32 Word counts: 10 words (title); 147 words (Abstract); 6004 words (Main Text) Tables: 3; Figure: 1; Supplementary figure: 1 Tamada & Kondo - 2 Abstract About 20 species of viruses belonging to five genera, Benyvirus, Furovirus, Pecluvirus, Pomovirus and Bymovirus, are known to be transmitted by plasmodiophorids. These viruses have all positive-sense single-stranded RNA genomes that consist of two to five RNA components. Three species of plasmodiophorids are recognized as vectors: Polymyxa graminis, P. betae, and Spongospora subterranea. The viruses can survive in soil within the long-lived resting spores of the vector. There are biological and genetic variations in both virus and vector species. Many of the viruses have become the causal agents of important diseases in major crops, such as rice, wheat, barley, rye, sugar beet, potato, and groundnut. Control measure is dependent on the development of the resistant cultivars. During the last half a century, several virus diseases have been rapidly spread and distributed worldwide. For the six major virus diseases, their geographical distribution, diversity, and genetic resistance are addressed. -

Viruses Virus Diseases Poaceae(Gramineae)
Viruses and virus diseases of Poaceae (Gramineae) Viruses The Poaceae are one of the most important plant families in terms of the number of species, worldwide distribution, ecosystems and as ingredients of human and animal food. It is not surprising that they support many parasites including and more than 100 severely pathogenic virus species, of which new ones are being virus diseases regularly described. This book results from the contributions of 150 well-known specialists and presents of for the first time an in-depth look at all the viruses (including the retrotransposons) Poaceae(Gramineae) infesting one plant family. Ta xonomic and agronomic descriptions of the Poaceae are presented, followed by data on molecular and biological characteristics of the viruses and descriptions up to species level. Virus diseases of field grasses (barley, maize, rice, rye, sorghum, sugarcane, triticale and wheats), forage, ornamental, aromatic, wild and lawn Gramineae are largely described and illustrated (32 colour plates). A detailed index Sciences de la vie e) of viruses and taxonomic lists will help readers in their search for information. Foreworded by Marc Van Regenmortel, this book is essential for anyone with an interest in plant pathology especially plant virology, entomology, breeding minea and forecasting. Agronomists will also find this book invaluable. ra The book was coordinated by Hervé Lapierre, previously a researcher at the Institut H. Lapierre, P.-A. Signoret, editors National de la Recherche Agronomique (Versailles-France) and Pierre A. Signoret emeritus eae (G professor and formerly head of the plant pathology department at Ecole Nationale Supérieure ac Agronomique (Montpellier-France). Both have worked from the late 1960’s on virus diseases Po of Poaceae . -

ICTV Code Assigned: 2011.001Ag Officers)
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2011.001aG officers) Short title: Change existing virus species names to non-Latinized binomials (e.g. 6 new species in the genus Zetavirus) Modules attached 1 2 3 4 5 (modules 1 and 9 are required) 6 7 8 9 Author(s) with e-mail address(es) of the proposer: Van Regenmortel Marc, [email protected] Burke Donald, [email protected] Calisher Charles, [email protected] Dietzgen Ralf, [email protected] Fauquet Claude, [email protected] Ghabrial Said, [email protected] Jahrling Peter, [email protected] Johnson Karl, [email protected] Holbrook Michael, [email protected] Horzinek Marian, [email protected] Keil Guenther, [email protected] Kuhn Jens, [email protected] Mahy Brian, [email protected] Martelli Giovanni, [email protected] Pringle Craig, [email protected] Rybicki Ed, [email protected] Skern Tim, [email protected] Tesh Robert, [email protected] Wahl-Jensen Victoria, [email protected] Walker Peter, [email protected] Weaver Scott, [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . -

Tamada-Text R3 HK-TT
Tamada & Kondo - 1 Journal of General Plant Pathology Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors Tetsuo Tamada* and Hideki Kondo Institute of Plant Science and Resources (IPSR), Okayama University, Kurashiki 710-0046, Japan. Current address for T. Tamada: Kita 10, Nishi 1, 13-2-606, Sapporo, 001-0010, Japan. *Corresponding author: Tetsuo Tamada; E-mail: [email protected] Total text pages 32 Word counts: 10 words (title); 147 words (Abstract); 6004 words (Main Text) Tables: 3; Figure: 1; Supplementary figure: 1 Tamada & Kondo - 2 Abstract About 20 species of viruses belonging to five genera, Benyvirus, Furovirus, Pecluvirus, Pomovirus and Bymovirus, are known to be transmitted by plasmodiophorids. These viruses have all positive-sense single-stranded RNA genomes that consist of two to five RNA components. Three species of plasmodiophorids are recognized as vectors: Polymyxa graminis, P. betae, and Spongospora subterranea. The viruses can survive in soil within the long-lived resting spores of the vector. There are biological and genetic variations in both virus and vector species. Many of the viruses have become the causal agents of important diseases in major crops, such as rice, wheat, barley, rye, sugar beet, potato, and groundnut. Control measure is dependent on the development of the resistant cultivars. During the last half a century, several virus diseases have been rapidly spread and distributed worldwide. For the six major virus diseases, their geographical distribution, diversity, and genetic resistance are addressed. Keywords Soil-borne viruses; Benyvirus; Furovirus; Pecluvirus; Pomovirus; Bymovirus; Vector transmission; Plasmodiophorids; Polymyxa; Spongospora Introduction There is a group of viruses to be transmitted by olpidium and plasmodiophorid vectors, which is called soil-borne fungus-transmitted viruses. -

Small Hydrophobic Viral Proteins Involved in Intercellular Movement of Diverse Plant Virus Genomes Sergey Y
AIMS Microbiology, 6(3): 305–329. DOI: 10.3934/microbiol.2020019 Received: 23 July 2020 Accepted: 13 September 2020 Published: 21 September 2020 http://www.aimspress.com/journal/microbiology Review Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes Sergey Y. Morozov1,2,* and Andrey G. Solovyev1,2,3 1 A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, Moscow, Russia 2 Department of Virology, Biological Faculty, Moscow State University, Moscow, Russia 3 Institute of Molecular Medicine, Sechenov First Moscow State Medical University, Moscow, Russia * Correspondence: E-mail: [email protected]; Tel: +74959393198. Abstract: Most plant viruses code for movement proteins (MPs) targeting plasmodesmata to enable cell-to-cell and systemic spread in infected plants. Small membrane-embedded MPs have been first identified in two viral transport gene modules, triple gene block (TGB) coding for an RNA-binding helicase TGB1 and two small hydrophobic proteins TGB2 and TGB3 and double gene block (DGB) encoding two small polypeptides representing an RNA-binding protein and a membrane protein. These findings indicated that movement gene modules composed of two or more cistrons may encode the nucleic acid-binding protein and at least one membrane-bound movement protein. The same rule was revealed for small DNA-containing plant viruses, namely, viruses belonging to genus Mastrevirus (family Geminiviridae) and the family Nanoviridae. In multi-component transport modules the nucleic acid-binding MP can be viral capsid protein(s), as in RNA-containing viruses of the families Closteroviridae and Potyviridae. However, membrane proteins are always found among MPs of these multicomponent viral transport systems. -
Us 2012/0074.014 A1 2 .. 1
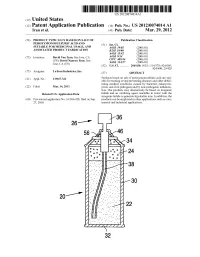
US 2012O074O14A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0074.014 A1 Tran et al. (43) Pub. Date: Mar. 29, 2012 (54) PRODUCT TYPICALLY BASED ON SALT OF Publication Classification PEROXYMONOSULFURIC ACID AND (51) Int. Cl SUITABLE FOR MEDICINAL USAGE, AND iBio/02 (2006.01) ASSOCATED PRODUCT FABRICATION B23P 19/00 (2006.01) A633/42 (2006.01) (75) Inventors: David Van Tran, San Jose, CA A6IR 9/14 (2006.01) (US); David Nguyen Tran, San CD7C 409/244 (2006.01) Jose, CA (US) A6II 3/327 (2006.01) s (52) U.S. Cl. ............. 206/438: 562/1; 514/578; 424/605; 424/400; 29/428 (73) Assignee: LuTran Industries, Inc. (57) ABSTRACT 21) Appl. No.: 13AO47,742 Products based on salt of peroxyperoxVmonosulfuric acid are suit (21) Appl. No 9 able for treating or/and preventing diseases and other debili tating medical conditions caused by bacterial, eukaryotic, (22) Filed: Mar. 14, 2011 prion, and viral pathogens and by non-pathogenic inflamma tion. The products may alternatively be based on inorganic Related U.S. Application Data halide and an oxidizing agent reactable in water with the inorganic halide to generate hypohalite ions. In addition, the (60) Provisional application No. 61/386,928, filed on Sep. products can be employed in other applications such as com 27, 2010. mercial and industrial applications. 5T. 2 .. 2. 1.4. J., Patent Application Publication Mar. 29, 2012 Sheet 1 of 2 US 2012/0074014 A1 cro Q-D 22 Fig. 2a Fig.2b Patent Application Publication Mar. 29, 2012 Sheet 2 of 2 US 2012/0074014 A1 90 92 US 2012/0074014 A1 Mar. -

Evidence to Support Safe Return to Clinical Practice by Oral Health Professionals in Canada During the COVID-19 Pandemic: a Repo
Evidence to support safe return to clinical practice by oral health professionals in Canada during the COVID-19 pandemic: A report prepared for the Office of the Chief Dental Officer of Canada. November 2020 update This evidence synthesis was prepared for the Office of the Chief Dental Officer, based on a comprehensive review under contract by the following: Paul Allison, Faculty of Dentistry, McGill University Raphael Freitas de Souza, Faculty of Dentistry, McGill University Lilian Aboud, Faculty of Dentistry, McGill University Martin Morris, Library, McGill University November 30th, 2020 1 Contents Page Introduction 3 Project goal and specific objectives 3 Methods used to identify and include relevant literature 4 Report structure 5 Summary of update report 5 Report results a) Which patients are at greater risk of the consequences of COVID-19 and so 7 consideration should be given to delaying elective in-person oral health care? b) What are the signs and symptoms of COVID-19 that oral health professionals 9 should screen for prior to providing in-person health care? c) What evidence exists to support patient scheduling, waiting and other non- treatment management measures for in-person oral health care? 10 d) What evidence exists to support the use of various forms of personal protective equipment (PPE) while providing in-person oral health care? 13 e) What evidence exists to support the decontamination and re-use of PPE? 15 f) What evidence exists concerning the provision of aerosol-generating 16 procedures (AGP) as part of in-person -

Armillaria Root Rot Fungi Host Single-Stranded RNA Viruses
www.nature.com/scientificreports OPEN Armillaria root rot fungi host single‑stranded RNA viruses Riikka Linnakoski1,5, Suvi Sutela1,5, Martin P. A. Coetzee2, Tuan A. Duong2, Igor N. Pavlov3,4, Yulia A. Litovka3,4, Jarkko Hantula1, Brenda D. Wingfeld2 & Eeva J. Vainio1* Species of Armillaria are distributed globally and include some of the most important pathogens of forest and ornamental trees. Some of them form large long‑living clones that are considered as one of the largest organisms on earth and are capable of long‑range spore‑mediated transfer as well as vegetative spread by drought‑resistant hyphal cords called rhizomorphs. However, the virus community infecting these species has remained unknown. In this study we used dsRNA screening and high‑throughput sequencing to search for possible virus infections in a collection of Armillaria isolates representing three diferent species: Armillaria mellea from South Africa, A. borealis from Finland and Russia (Siberia) and A. cepistipes from Finland. Our analysis revealed the presence of both negative‑ sense RNA viruses and positive‑sense RNA viruses, while no dsRNA viruses were detected. The viruses included putative new members of virus families Mymonaviridae, Botourmiaviridae and Virgaviridae and members of a recently discovered virus group tentatively named “ambiviruses” with ambisense bicistronic genomic organization. We demonstrated that Armillaria isolates can be cured of viruses by thermal treatment, which enables the examination of virus efects on host growth and phenotype using isogenic virus‑infected and virus‑free strains. Te fungal genus Armillaria (Fr.) Staude includes more than 40 described species1. Tey are mainly known as notorious plant pathogens of managed natural forests and plantations of non-native tree species that infect hundreds of diferent plants, including economically important conifers (e.g. -

RNA/RNA Interactions Involved in the Regulation of Benyviridae Viral Cicle Mattia Dall’Ara
RNA/RNA interactions involved in the regulation of Benyviridae viral cicle Mattia Dall’Ara To cite this version: Mattia Dall’Ara. RNA/RNA interactions involved in the regulation of Benyviridae viral cicle. Vegetal Biology. Université de Strasbourg; Università degli studi (Bologne, Italie), 2018. English. NNT : 2018STRAJ019. tel-02003448 HAL Id: tel-02003448 https://tel.archives-ouvertes.fr/tel-02003448 Submitted on 1 Feb 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université de Strasbourg et Università di Bologna Thèse en co-tutelle Présentée à la FACULTÉ DES SCIENCES DE LA VIE En vue de l'obtention du titre de DOCTEUR DE L'UNIVERSITÉ DE STRASBOURG Discipline : Sciences de la Vie et de la Santé, Spécialité : Aspects moléculaires et cellulaires de la biologie Par Mattia Dall’Ara RNA/RNA interactions involved in the regulation of Benyviridae viral cicle Soutenue publiquement le 18 mai 2018 devant la Commission d'Examen : Dr. Stéphane BLANC Rapporteur Externe Dr. Renato BRANDIMARTI Rapporteur Externe Dr. Roland MARQUET Examinateur Interne Dr. Mirco IOTTI Examinateur Interne Pr. David GILMER Directeur de Thèse Dr. Claudio RATTI Co-directeur de Thèse DISTAL Dipartimento di Scienze e Tecnologie Agro-Alimentari Bologne Italie Acknowledgments First of all I want to thank my tutors David and Claudio. -

Multipartite Viruses: Organization, Emergence and Evolution
UNIVERSIDAD AUTÓNOMA DE MADRID FACULTAD DE CIENCIAS DEPARTAMENTO DE BIOLOGÍA MOLECULAR MULTIPARTITE VIRUSES: ORGANIZATION, EMERGENCE AND EVOLUTION TESIS DOCTORAL Adriana Lucía Sanz García Madrid, 2019 MULTIPARTITE VIRUSES Organization, emergence and evolution TESIS DOCTORAL Memoria presentada por Adriana Luc´ıa Sanz Garc´ıa Licenciada en Bioqu´ımica por la Universidad Autonoma´ de Madrid Supervisada por Dra. Susanna Manrubia Cuevas Centro Nacional de Biotecnolog´ıa (CSIC) Memoria presentada para optar al grado de Doctor en Biociencias Moleculares Facultad de Ciencias Departamento de Biolog´ıa Molecular Universidad Autonoma´ de Madrid Madrid, 2019 Tesis doctoral Multipartite viruses: Organization, emergence and evolution, 2019, Madrid, Espana. Memoria presentada por Adriana Luc´ıa-Sanz, licenciada en Bioqumica´ y con un master´ en Biof´ısica en la Universidad Autonoma´ de Madrid para optar al grado de doctor en Biociencias Moleculares del departamento de Biolog´ıa Molecular en la facultad de Ciencias de la Universidad Autonoma´ de Madrid Supervisora de tesis: Dr. Susanna Manrubia Cuevas. Investigadora Cient´ıfica en el Centro Nacional de Biotecnolog´ıa (CSIC), C/ Darwin 3, 28049 Madrid, Espana. to the reader CONTENTS Acknowledgments xi Resumen xiii Abstract xv Introduction xvii I.1 What is a virus? xvii I.2 What is a multipartite virus? xix I.3 The multipartite lifecycle xx I.4 Overview of this thesis xxv PART I OBJECTIVES PART II METHODOLOGY 0.5 Database management for constructing the multipartite and segmented datasets 3 0.6 Analytical -

University of Oklahoma Graduate College Viral
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE VIRAL METAGENOMICS AND ANTHROPOLOGY IN THE AMERICAS A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By ANDREW T. OZGA Norman, Oklahoma 2015 VIRAL METAGENOMICS AND ANTHROPOLOGY IN THE AMERICAS A DISSERTATION APPROVED FOR THE DEPARTMENT OF ANTHROPOLOGY BY ______________________________ Dr. Cecil M. Lewis, Jr., Chair ______________________________ Dr. Katherine Hirschfeld ______________________________ Dr. Paul Spicer ______________________________ Dr. Kermyt G. Anderson ______________________________ Dr. Tyrrell Conway © Copyright by ANDREW T. OZGA 2015 All Rights Reserved. Acknowledgements This dissertation would not have been possible without the guidance and encouragement of a number of professors, colleagues, friends, and family members. First and foremost I’d like to thank my advisor and chair, Dr. Cecil Lewis. His guidance and critiques throughout the project design, execution, and drafting process were invaluable to my completion of this degree. I’d also like to thank my doctoral committee: Dr. Tassie Hirschfeld, Dr. KG Anderson, Dr. Paul Spicer, and Dr. Tyrrell Conway for their support and insight regarding my project design and manuscript draft. I must thank those that graciously provided fecal samples for this project including the Cheyenne & Arapaho Nation, University of Oklahoma colleagues and students, and those from the coastal and jungle regions of Peru. Additionally, I’d like to thank my LMAMR colleagues including Dr. Christina Warinner and Dr. Jiawu Xu for their encouragement and valuable knowledge. This includes Dr. Alexandra Obregon-Tito and Raul Tito for their relentless support of my pursuit of a Ph.D., their assistance in sample processing, and their assistance in the Peruvian IRB process and sample collection. -

Plant Virus RNA Replication
eLS Plant Virus RNA Replication Alberto Carbonell*, Juan Antonio García, Carmen Simón-Mateo and Carmen Hernández *Corresponding author: Alberto Carbonell ([email protected]) A22338 Author Names and Affiliations Alberto Carbonell, Instituto de Biología Molecular y Celular de Plantas (CSIC-UPV), Campus UPV, Valencia, Spain Juan Antonio García, Centro Nacional de Biotecnología (CSIC), Madrid, Spain Carmen Simón-Mateo, Centro Nacional de Biotecnología (CSIC), Madrid, Spain Carmen Hernández, Instituto de Biología Molecular y Celular de Plantas (CSIC-UPV), Campus UPV, Valencia, Spain *Advanced article Article Contents • Introduction • Replication cycles and sites of replication of plant RNA viruses • Structure and dynamics of viral replication complexes • Viral proteins involved in plant virus RNA replication • Host proteins involved in plant virus RNA replication • Functions of viral RNA in genome replication • Concluding remarks Abstract Plant RNA viruses are obligate intracellular parasites with single-stranded (ss) or double- stranded RNA genome(s) generally encapsidated but rarely enveloped. For viruses with ssRNA genomes, the polarity of the infectious RNA (positive or negative) and the presence of one or more genomic RNA segments are the features that mostly determine the molecular mechanisms governing the replication process. RNA viruses cannot penetrate plant cell walls unaided, and must enter the cellular cytoplasm through mechanically-induced wounds or assisted by a 1 biological vector. After desencapsidation, their genome remains in the cytoplasm where it is translated, replicated, and encapsidated in a coupled manner. Replication occurs in large viral replication complexes (VRCs), tethered to modified membranes of cellular organelles and composed by the viral RNA templates and by viral and host proteins.