Investigations of CHO 1-15 and Silk Gland Cell Line Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fall ’17 Footwear Trends Smart Textiles Tech Advances Trade Show Previews & Recaps Making It in America
TEXTILE INSIGHT Trends In Apparel & Footwear Design and Innovation • January/February 2017 iNvEnToLoGy HOW THE TEXTILE INDUSTRY IS INVENTING ITS FUTURE Fall ’17 Footwear Trends Smart Textiles Tech Advances Trade Show Previews & Recaps Making it in America A FORMULA4 MEDIA PUBLICATION TEXTILEINSIGHT.COM A breakthrough fiber innovation you have to feel to believe. Eastman Avra performance fibers wick better, dry faster, and keep you at your coolest and most comfortable. AVRAfromEastman.com ©2017 Eastman Chemical Company. Eastman brands referenced herein are trademarks of Eastman Chemical Company or one of its subsidiaries. The ® used on Eastman brands denotes registered trademark status in the U.S.; marks may also be registered internationally. JANUARY/FEBRUARY 2017 JANUARY/FEBRUARY Executive Editor Mark Sullivan [email protected] 646-319-7878 Editor /Associate Publisher Emily Walzer [email protected] Managing Editor Cara Griffin Art Director Francis Klaess Associate Art Director Mary McGann Contributing Editors Suzanne Blecher Kurt Gray Jennifer Ernst Beaudry Kathlyn Swantko Publisher Jeff Nott [email protected] 516-305-4711 Advertising Jeff Gruenhut [email protected] 404-467-9980 Christina Henderson 516-305-4710 [email protected] Troy Leonard [email protected] 352-624-1561 Katie O’Donohue [email protected] 828-244-3043 Sam Selvaggio [email protected] 212-398-5021 Production Brandon Christie 516 305-4710 [email protected] Business Manager Marianna Rukhvarger 516-305-4709 [email protected] Biosteel is the latest knit upper shoe technology from Adidas set to debut this year. Subscriptions store.formula4media.com Formula4 Media Publications 6 / In the Market 40 / Made in America Sports Insight The New Year begins with trade show previews, reports on Before “Think locally, act globally” became a popular buzz- Outdoor Insight Footwear Insight Performance Days, advances in digital textiles printing, and phrase, IDEAL Fastener was putting the strategy to work. -

Spider Silk for Tissue Engineering Applications
molecules Review Spider Silk for Tissue Engineering Applications Sahar Salehi 1, Kim Koeck 1 and Thomas Scheibel 1,2,3,4,5,* 1 Department for Biomaterials, University of Bayreuth, Prof.-Rüdiger-Bormann-Strasse 1, 95447 Bayreuth, Germany; [email protected] (S.S.); [email protected] (K.K.) 2 The Bayreuth Center for Colloids and Interfaces (BZKG), University of Bayreuth, Universitätsstraße 30, 95447 Bayreuth, Germany 3 The Bayreuth Center for Molecular Biosciences (BZMB), University of Bayreuth, Universitätsstraße 30, 95447 Bayreuth, Germany 4 The Bayreuth Materials Center (BayMAT), University of Bayreuth, Universitätsstraße 30, 95447 Bayreuth, Germany 5 Bavarian Polymer Institute (BPI), University of Bayreuth, Universitätsstraße 30, 95447 Bayreuth, Germany * Correspondence: [email protected]; Tel.: +49-0-921-55-6700 Received: 15 January 2020; Accepted: 6 February 2020; Published: 8 February 2020 Abstract: Due to its properties, such as biodegradability, low density, excellent biocompatibility and unique mechanics, spider silk has been used as a natural biomaterial for a myriad of applications. First clinical applications of spider silk as suture material go back to the 18th century. Nowadays, since natural production using spiders is limited due to problems with farming spiders, recombinant production of spider silk proteins seems to be the best way to produce material in sufficient quantities. The availability of recombinantly produced spider silk proteins, as well as their good processability has opened the path towards modern biomedical applications. Here, we highlight the research on spider silk-based materials in the field of tissue engineering and summarize various two-dimensional (2D) and three-dimensional (3D) scaffolds made of spider silk. -

Spidroin Striped Micropattern Promotes Chondrogenic Differentiation of Human Wharton's Jelly Mesenchymal Stem Cells
Spidroin Striped Micropattern Promotes Chondrogenic Differentiation of Human Wharton’s Jelly Mesenchymal Stem Cells Anggraini Barlian ( [email protected] ) Bandung Institute of Technology Dinda Hani’ah Arum Saputri Bandung Institute of Technology Adriel Hernando Bandung Institute of Technology Ekavianty Prajatelistia Bandung Institute of Technology Hutomo Tanoto Bandung Institute of Technology Research Article Keywords: chondrogenesis, human Wharton’s jelly, mesenchymal stem cells, microcontact printing, spidroin Posted Date: April 23rd, 2021 DOI: https://doi.org/10.21203/rs.3.rs-445399/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Spidroin Striped Micropattern Promotes Chondrogenic Differentiation of Human Wharton’s Jelly Mesenchymal Stem Cells Anggraini Barlian*1,2, Dinda Hani’ah Arum Saputri1, Adriel Hernando1, Ekavianty Prajatelistia3, Hutomo Tanoto3 1School of Life Sciences and Technology, Bandung Institute of Technology, Bandung, West Java, 40132, Indonesia 2Research Center for Nanosciences and Nanotechnology, Bandung Institute of Technology, Bandung, West Java, 40132, Indonesia 3Faculty of Mechanical and Aerospace Engineering, Bandung Institute of Technology, Bandung, West Java, 40132, Indonesia *Corresponding author email: [email protected] ABSTRACT Cartilage tissue engineering, particularly micropattern, can influence the biophysical properties of mesenchymal stem cells (MSCs) leading to chondrogenesis. In this research, human Wharton’s jelly MSCs (hWJ-MSCs) were grown on a striped micropattern containing spider silk protein (spidroin) from Argiope appensa. This research aims to direct hWJ-MSCs chondrogenesis using micropattern made of spidroin bioink as opposed to fibronectin that often used as the gold standard. Cells were cultured on striped micropattern of 500 µm and 1000 µm width sizes without chondrogenic differentiation medium for 21 days. -

To Be Published in Gazette of India, Extra Ordinary, Part 1, Section1
To be published in Gazette of India, Extra ordinary, Part 1, Section1 F. No. 15/16/2013-DGAD Government of India Ministry of Commerce & Industry Department of Commerce (Directorate General of Anti Dumping & Allied Duties) 4th Floor, Jeevan Tara Building, 5, Parliament Street, New Delhi 110001 Dated the 23rd March, 2015 NOTIFICATION (Final Findings) Subject: Final Findings in the sunset review of anti-dumping duty imposed on the imports of Acrylic Fibre originating in or exported from Korea RP and Thailand-reg. A. BACKGROUND OF THE CASE 1. No. 15/16/2013-DGAD:- Whereas having regard to the Customs Tariff Act, 1975 as amended from time to time (herein after referred to as the Act) and the Customs Tariff (Identification, Assessment and Collection of Duty on Dumped Articles and for Determination of Injury) Rules, 1995 (herein after referred to as the AD Rules), the Designated Authority (herein after referred to as the Authority), initiated the original anti dumping investigation in respect of the imports of Acrylic Fibre (hereinafter referred to as the subject goods) originating in or exported from USA, Thailand and Korea RP on 13.9.1996 and definitive anti dumping duty was recommended vide Final Findings Notification No. 47/ADD/1W dated 14.10.1997. The Central Government had imposed the anti dumping duty. The sunset review of the anti dumping duty so imposed against USA, Thailand and Korea RP was initiated by the Authority vide Notification No. 26/1/2001-DGAD dated 07.08.2001 and the Final Findings were issued vide Notification No. 26/1/2001-DGAD dated 06.08.2002. -

U.S. EPA, Pesticide Product Label, CUTTER INSECT REPELLENT
U.S. ENVIRONMENTAL PROTECTION AGENCY EPA Reg. Date of Issuance: Office of Pesticide Programs Number: Registration Division (H750SC) 401 "M" St., S.W. 121-91 Washington, D.C. 20460 OCT 2 a 2005 Term of Issuance: NOTICE OF PESTICIDE: Conditional _x__ Registration Reregistration Name of Pesticide Product: (under FIFRA. as amended) CUTTER Insect Repellent 15KP Name and Address of Registrant (include ZIP Code): Spectrum Division of United Industries P.O. Box 142642 St. Louis, MO 63114-0642 Note: Changes in labeling differing in substance from that accepted in connection with this registration must be submitted to and accepted by the Registration Division prior to use of the label in commerce. In any correspondence on this product always refer to the above EPA registration number. On the basis of information furnished by the registrant, the above named pesticide is hereby registered/reregistered under the Federal Insecticide, Fungicide and Rodenticide Act. Registration is in no way to be construed as an endorsement or recommendation of this product by the Agency. In order to protect health and the environment, the Administrator, on his motion, may at any time suspend or cancel the registration of a pesticide in accordance with the Act. The acceptance of any name in connection with the registration of a product under this Act is not to be construed as giving the registrant a right to exclusive use of the name or to its use if it has been covered by others. This product is conditionally registered in accordance with FIFRA sec. 3(c) (7) (A) provided that you: 1. -

Tarantulas and Social Spiders
Tarantulas and Social Spiders: A Tale of Sex and Silk by Jonathan Bull BSc (Hons) MSc ICL Thesis Presented to the Institute of Biology of The University of Nottingham in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy The University of Nottingham May 2012 DEDICATION To my parents… …because they both said to dedicate it to the other… I dedicate it to both ii ACKNOWLEDGEMENTS First and foremost I would like to thank my supervisor Dr Sara Goodacre for her guidance and support. I am also hugely endebted to Dr Keith Spriggs who became my mentor in the field of RNA and without whom my understanding of the field would have been but a fraction of what it is now. Particular thanks go to Professor John Brookfield, an expert in the field of biological statistics and data retrieval. Likewise with Dr Susan Liddell for her proteomics assistance, a truly remarkable individual on par with Professor Brookfield in being able to simplify even the most complex techniques and analyses. Finally, I would really like to thank Janet Beccaloni for her time and resources at the Natural History Museum, London, permitting me access to the collections therein; ten years on and still a delight. Finally, amongst the greats, Alexander ‘Sasha’ Kondrashov… a true inspiration. I would also like to express my gratitude to those who, although may not have directly contributed, should not be forgotten due to their continued assistance and considerate nature: Dr Chris Wade (five straight hours of help was not uncommon!), Sue Buxton (direct to my bench creepy crawlies), Sheila Keeble (ventures and cleans where others dare not), Alice Young (read/checked my thesis and overcame her arachnophobia!) and all those in the Centre for Biomolecular Sciences. -
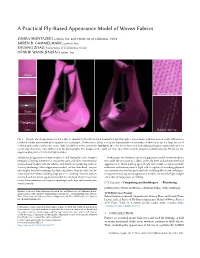
A Practical Ply-Based Appearance Model of Woven Fabrics
A Practical Ply-Based Appearance Model of Woven Fabrics ZAHRA MONTAZERI, Luxion, Inc. and University of California, Irvine SØREN B. GAMMELMARK, Luxion, Inc. SHUANG ZHAO, University of California, Irvine HENRIK WANN JENSEN, Luxion, Inc. Photo Ours Frontlit close-up Photo Ours Backlit close-up Fig. 1. Our ply-based appearance model is able to simulate both reflected and transmitted light through complex weave patterns as seen on the left andour rendered results match with photographs of a real sample. Furthermore, it has a compact representation that makes it efficient to use for large pieces of clothing such as the scarf shown on the right. In addition to the anisotropic highlights, the color due to front and back lighting changes considerably and our model reproduces the color shift seen in the photographs. The images on the right are close-ups of the scarf showing the individual yarns. Pleaseseethe supplementary video for the full light rotation. Simulating the appearance of woven fabrics is challenging due to the complex In this paper, we introduce a practical appearance model for woven fabrics. interplay of lighting between the constituent yarns and fibers. Conventional We model the structure of a fabric at the ply level and simulate the local surface-based models lack the fidelity and details for producing realistic appearance of fibers making up each ply. Our model accounts for both close-up renderings. Micro-appearance models, on the other hand, can pro- reflection and transmission of light and is capable of matching physical duce highly detailed renderings by depicting fabrics fiber-by-fiber, but be- measurements better than prior methods including fiber based techniques. -

Textile Design: a Suggested Program Guide
DOCUMENT RESUME CI 003 141 ED 102 409 95 Program Guide.Fashion TITLE Textile Design: A Suggested Industry Series No. 3. Fashion Inst. of Tech.,New York, N.T. INSTITUTION Education SPONS AGENCY Bureau of Adult,Vocational, and Technictl (DREW /OE), Washington,D.C. PUB DATE 73 in Fashion Industry NOTE 121p.; For other documents Series, see CB 003139-142 and CB 003 621 Printing AVAILABLE FROM Superintendent of Documents,U.S. Government Office, Washington, D.C.20402 EDRS PRICE NP -$0.76 HC-$5.70 PLUS POSTAGE Behavioral Objectives; DESCRIPTORS Adult, Vocational Education; Career Ladders; *CurriculumGuides; *Design; Design Crafts; EducationalEquipment; Employment Opportunities; InstructionalMaterials; *Job Training; Needle Trades;*Occupational Rome Economics; OccupationalInformation; Program Development; ResourceGuides; Resource Units; Secondary Education;Skill Development;*Textiles Instruction IDENTIFIERS *Fashion Industry ABSTRACT The textile designguide is the third of aseries of resource guidesencompassing the various five interrelated program guide is disensions of the fashionindustry. The job-preparatory conceived to provide youthand adults withintensive preparation for and also with careeradvancement initial entry esploysent jobs within the textile opportunities withinspecific categories of provides an overviewof the textiledesign field, industry. The guide required of workers. It occupational opportunities,and cospetencies contains outlines of areasof instruction whichinclude objectives to suggestions for learning be achieved,teaching -

Mussels As a Model System for Integrative Ecomechanics
UC Santa Barbara UC Santa Barbara Previously Published Works Title Mussels as a model system for integrative ecomechanics. Permalink https://escholarship.org/uc/item/0xr832ct Journal Annual review of marine science, 7(1) ISSN 1941-1405 Authors Carrington, Emily Waite, J Herbert Sarà, Gianluca et al. Publication Date 2015 DOI 10.1146/annurev-marine-010213-135049 Peer reviewed eScholarship.org Powered by the California Digital Library University of California MA07CH19-Carrington ARI 20 November 2014 8:4 Mussels as a Model System for Integrative Ecomechanics Emily Carrington,1 J. Herbert Waite,2 Gianluca Sara,` 3 and Kenneth P. Sebens1 1Department of Biology and Friday Harbor Laboratories, University of Washington, Friday Harbor, Washington 98250; email: [email protected], [email protected] 2Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara, California 93106; email: [email protected] 3Dipartimento di Scienze della Terra e del Mare, University of Palermo, 90128 Palermo, Italy; email: [email protected] Annu. Rev. Mar. Sci. 2015. 7:443–69 Keywords First published online as a Review in Advance on byssus, dislodgment, dynamic energy budget, fitness, mussel foot proteins, August 25, 2014 tenacity The Annual Review of Marine Science is online at marine.annualreviews.org Abstract This article’s doi: Mussels form dense aggregations that dominate temperate rocky shores, and 10.1146/annurev-marine-010213-135049 they are key aquaculture species worldwide. Coastal environments are dy- Copyright c 2015 by Annual Reviews. Annu. Rev. Marine. Sci. 2015.7:443-469. Downloaded from www.annualreviews.org namic across a broad range of spatial and temporal scales, and their changing All rights reserved abiotic conditions affect mussel populations in a variety of ways, including Access provided by University of California - Santa Barbara on 01/30/15. -

Cyromazine) During Woolscouring and Its Effects on the Aquatic Environment the Fate of Vetrazin® (Cyromazine) During
Lincoln University Digital Thesis Copyright Statement The digital copy of this thesis is protected by the Copyright Act 1994 (New Zealand). This thesis may be consulted by you, provided you comply with the provisions of the Act and the following conditions of use: you will use the copy only for the purposes of research or private study you will recognise the author's right to be identified as the author of the thesis and due acknowledgement will be made to the author where appropriate you will obtain the author's permission before publishing any material from the thesis. THE FATE OF VETRAZIN@ (CYROMAZINE) DURING WOOLSCOURING AND ITS EFFECTS ON THE AQUATIC ENVIRONMENT THE FATE OF VETRAZIN® (CYROMAZINE) DURING WOOLSCOURING AND ITS EFFECTS ON THE AQUATIC ENVIRONMENT A thesis submitted in fulfilment of the requirements for the degree of DOCTOR OF PHILOSOPHY in AQUATIC TOXICOLOGY at LINCOLN UNIVERSITY P.W. Robinson 1995 " ; " i Abstract of a thesis submitted in partial fulfllment of the requirement for the Degree of Doctor of Philosophy THE FATE OF VETRAZIN® (CYROMAZINE) DURING WOOLSCOURING AND ITS EFFECTS ON THE AQUATIC ENVIRONMENT by P.W. Robinson A number of ectoparasiticides are used on sheep to protect the animals from ill health associated with infestations of lice and the effects of fly-strike. Most of the compounds currently in use are organophosphate- or pyrethroid-based and have been used for 15-20 years, or more. In more recent times, as with other pest control strategies, there has been a tendency to introduce 'newer' pesticides, principally in the form of insect growth regulators (IGRs). -

Spinning and Applications of Spider Silk
Frontiers in Science 2012, 2(5): 92-100 DOI: 10.5923/j.fs.20120205.02 Spinning and Applications of Spider Silk Kunal Singha*, Subhankar Maity, Mrinal Singha Department of Textile Engineering, Panipat Institute of Engineering & Technology, Harayana, India Abstract Spider silk is specially produced from the different species (sp.) of spider throughout the globe, specially the most fine and mechanically sound silk produced from nephila sp. The fibriller parallel model of spider silk gives higher mechanical properties with better Young’s modulus and lower stress than normal silk coming from the silk-moth. In this review work we dicuss about the collection, composition, spinning and molecular structure, mechanical properties with its application of the spider silk. Ke ywo rds Spider Silk, Nephila Sp., Fibrille r Pa ralle l Model, Young’s Modulus, Spinning, Mechanical Properties 1. Introduction Spider silk refers to a wide range of continuous filaments 2. Macromolecular Structure of Silk spun by the several species of lepidoptera & arthropoda. Spidroin Spiders have six or seven sets of glands spun different fibers. Major ampullate dragline silk is comprised of two proteins There is different type of spider species; but the most joined together via 3-5 disulfide bonds near their common species of the spider which produced the spider C-termini[12]; the two proteins which are found in spider dragline silk is Araneus diadematus[1, 2]. silk are : spidroin 1 and spidroin 2. There are various types of spider species available in the nature and their habitat, location, life cycle are also totally 2.1. Secondary Structure of Dragline Silk different. -

Production and Purification of Synthetic Minor Ampullate Silk Proteins
Utah State University DigitalCommons@USU All Graduate Theses and Dissertations Graduate Studies 12-2018 Production and Purification of Synthetic Minor Ampullate Silk Proteins Danielle A. Gaztambide Utah State University Follow this and additional works at: https://digitalcommons.usu.edu/etd Part of the Engineering Commons Recommended Citation Gaztambide, Danielle A., "Production and Purification of Synthetic Minor Ampullate Silk Proteins" (2018). All Graduate Theses and Dissertations. 7306. https://digitalcommons.usu.edu/etd/7306 This Thesis is brought to you for free and open access by the Graduate Studies at DigitalCommons@USU. It has been accepted for inclusion in All Graduate Theses and Dissertations by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. PRODUCTION AND PURIFICATION OF SYNTHETIC MINOR AMPULLATE SILK PROTEINS by Danielle A. Gaztambide A thesis submitted in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE in Biological Engineering Approved: ______________________ ____________________ Randolph V. Lewis, Ph.D Jixun Zhan, Ph.D Major Professor Committee Member ______________________ ____________________ Ronald Sims, Ph.D Richard S. Inouye, Ph.D Committee Member School of Graduate Studies UTAH STATE UNIVERSITY Logan, Utah 2018 ii Copyright © Danielle A. Gaztambide 2018 All Rights Reserved iii ABSTRACT Production and Purification of Recombinant Minor Ampullate Silk Proteins by Danielle A. Gaztambide, Master of Science Utah State University, 2018 Major Professor: Randolph V. Lewis, Ph.D. Department: Biological Engineering Spider silks long have been recognized as some of nature’s most impressive materials. By divergent evolution, orb-weaving spiders produce and utilize six distinct silk fibers and one glue for a variety of biological roles.