Staphylococcus Spp. from Wild Mammals in Aragón (Spain): Antibiotic Resistance Status
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Appendix Lagomorph Species: Geographical Distribution and Conservation Status
Appendix Lagomorph Species: Geographical Distribution and Conservation Status PAULO C. ALVES1* AND KLAUS HACKLÄNDER2 Lagomorph taxonomy is traditionally controversy, and as a consequence the number of species varies according to different publications. Although this can be due to the conservative characteristic of some morphological and genetic traits, like general shape and number of chromosomes, the scarce knowledge on several species is probably the main reason for this controversy. Also, some species have been discovered only recently, and from others we miss any information since they have been first described (mainly in pikas). We struggled with this difficulty during the work on this book, and decide to include a list of lagomorph species (Table 1). As a reference, we used the recent list published by Hoffmann and Smith (2005) in the “Mammals of the world” (Wilson and Reeder, 2005). However, to make an updated list, we include some significant published data (Friedmann and Daly 2004) and the contribu- tions and comments of some lagomorph specialist, namely Andrew Smith, John Litvaitis, Terrence Robinson, Andrew Smith, Franz Suchentrunk, and from the Mexican lagomorph association, AMCELA. We also include sum- mary information about the geographical range of all species and the current IUCN conservation status. Inevitably, this list still contains some incorrect information. However, a permanently updated lagomorph list will be pro- vided via the World Lagomorph Society (www.worldlagomorphsociety.org). 1 CIBIO, Centro de Investigaça˜o em Biodiversidade e Recursos Genéticos and Faculdade de Ciˆencias, Universidade do Porto, Campus Agrário de Vaira˜o 4485-661 – Vaira˜o, Portugal 2 Institute of Wildlife Biology and Game Management, University of Natural Resources and Applied Life Sciences, Gregor-Mendel-Str. -

Espécies Potencialmente Ameaçadas 2019 VF4 Pivott.Xlsx
List of Potentially Impacted Species by EDP activities Legenda (1) threat levels: i. Critically endangered (CR); ii. Endangered (EN); iii. Vulnerable (VU); iv. Near threatened (NT) and v. Least concern (LC) 31/12/2019 Species IUCN Red List of Threatened Species (1) National Red List Occurrence Scientific name Common name (EN) Common name (country of occurrence) Reino/Kingdom Classe/Class 2020‐01 latest assessment National Red List Country Tecnology Acanthochelys spixii Black Spiny‐necked Swamp Turtle cágado‐feio Animalia Reptilia NT 1‐ago‐06 Brazil Windpower Acanthodactylus erythrurus Animalia Reptilia LC 14‐dez‐08 Portugal Windpower Spiny‐footed Lizard Lagartixa‐de‐dedos‐denteados NT Accipiter gentilis Northern Goshawk Açor Animalia Aves LC 31‐mar‐15 Portugal Windpower VU Accipiter gentilis Northern Goshawk Azor común Animalia Aves LC 31‐mar‐15 NE Spain Windpower Accipiter gentilis Animalia Aves LC 31‐mar‐15 n/a Poland Windpower Northern Goshawk Jastrząb zwyczajny Accipiter gentilis Northern Goshawk Azor común Animalia Aves LC 31‐mar‐15 NE Spain Transmision grid‐ Renew Accipiter gentilis Northern Goshawk Azor Aves LC 31‐mar‐15 Spain Solar FV Animalia NT Accipiter nisus Eurasian sparrowhawk Gavião Animalia Aves LC 1‐out‐16 Portugal Windpower LC Accipiter nisus Eurasian sparrowhawk Gavilán común Animalia Aves LC 1‐out‐16 NE Spain Windpower Accipiter nisus Eurasian sparrowhawk Eurasian sparrowhawk Animalia Aves LC 1‐out‐16 France Windpower LC Eurasian Sparrowhawk Eurasian Sparrowhawk Accipiter nisus Animalia Aves LC 1‐out‐16 Italy Windpower -

Hare Quiz: Questions and Answers
kupidonia.com Hare Quiz: questions and answers Hare Quiz: questions and answers - 1 / 4 kupidonia.com 1. Which animal family does hair belong to? Ochotonidae Lagomorpha Leporidae 2. Which subgenus is Granada hare belong to? Lepus Eulagos Indolagus 3. Which species does not belong to Subgenus Indolagus? Indian hare Chinese hare Hainan hare 4. What is a baby hair called? A leveret A jackrabbits A leporidus 5. What is a group of hairs called? A pack A legion Hare Quiz: questions and answers - 2 / 4 kupidonia.com A drove 6. How many species of hare exist? 34 32 28 7. According to African folktales, what is a hare known as? A moon creature An unfaithful lover A trickster 8. How many chromosomes do hares have? 44 46 48 9. What is the scientific name for Snowshoe hare? Lepus americanus Lepus alleni Lepus castroviejoi 10. What is the name of the German dish that is made of marinated hare? Jugged hare Lagos Stifado Hasenpfeffer Hare Quiz: questions and answers - 3 / 4 kupidonia.com Hare Quiz: questions and answers Right answers 1. Which animal family does hair belong to? Leporidae 2. Which subgenus is Granada hare belong to? Eulagos 3. Which species does not belong to Subgenus Indolagus? Chinese hare 4. What is a baby hair called? A leveret 5. What is a group of hairs called? A drove 6. How many species of hare exist? 32 7. According to African folktales, what is a hare known as? A trickster 8. How many chromosomes do hares have? 48 9. What is the scientific name for Snowshoe hare? Lepus americanus 10. -

May 16, 2011 Federal Trade Commission Office of the Secretary
May 16, 2011 Federal Trade Commission Office of the Secretary, Room H–113 (Annex O) 600 Pennsylvania Avenue, NW Washington, DC 20580 RE: Advance Notice of Proposed Rulemaking under the Fur Products Labeling Act; Matter No. P074201 On behalf of the more than 11 million members and supporters of The Humane Society of the United States (HSUS), I submit the following comments to be considered regarding the Federal Trade Commission’s (FTC) advance notice of proposed rulemaking under the federal Fur Products Labeling Act (FPLA), 16 U.S.C. § 69, et seq. The rulemaking is being proposed in response to the Truth in Fur Labeling Act (TFLA), Public Law 111–113, enacted in December 2010, which eliminates the de minimis value exemption from the FPLA, 16 U.S.C. § 69(d), and directs the FTC to initiate a review of the Fur Products Name Guide, 16 C.F.R. 301.0. Thus, the FTC indicated in its notice that it is specifically seeking comment on the Name Guide, though the agency is also generally seeking comment on its fur rules in their entirety. As discussed below, the HSUS believes that there is a continuing need for the fur rules and for more active enforcement of these rules by the FTC. The purpose of the FPLA and the fur rules is to ensure that consumers receive truthful and accurate information about the fur content of the products they are purchasing. Unfortunately, sales of unlabeled and mislabeled fur garments, and inaccurate or misleading advertising of fur garments, remain all too common occurrences in today’s marketplace. -

Klasse Orde Familie Geslacht Soort Ondersoort Vertaling Engels Frans
Blad1 A B C D E F G H I J K L 1 Klasse Orde Familie Geslacht Soort Ondersoort Vertaling Engels Frans Duits Spaans Nederlands 2 Mammalia Zoogdieren Mammals Mammif ères Säugetiere Mamíferos Zoogdieren 3 Lagomorpha Haasachtigen Lagomorphs Lagomorphes Hasenartige Lagomorfos Haasachtigen 4 Ochotonidae van Ochotona Pikas Ochotone s Pfeifhasen Ochotónidos Fluithazen 5 Ochotona uit het Mongools Pikas Pikas Pfeifhasen Picas Fluithazen 6 O. dauurica Dauria (Rusland) Daurian pika Pika de daourie Daurien-Pfeifhase Pica de Dauria Daurische fluithaas 7 O.d. dauurica Eastern Daurian pika Oostdaurische fluithaas 8 O.d. huangensis Huang He rivier Quinghai Daurian pika Qinghaifluithaas 9 O.d. latibullata grote blaas Uvs Nuur Daurian pika Uvs Nuurfluithaas 10 O. thibetana Tibetaans Moupin pika Pika du Moupin Tibet-Pfeifhase Pica de Moupin Tibetaanse fluithaas 11 O.t. thibetana Forest pika Woudfluithaas 12 O.t. nanggenica Nanggen, China Nanggen pika Nanggenfluithaas 13 O.T. sikimaria Sikkim Sikkim pika Sikkimfluithaas 14 O. syrinx Naam van een nimf Tsing-ling pika Pika des Quinglin Tsing-Ling-Pfeifhase Pica de Tsing-ling Tsing-lingfluithaas 15 O.s. syrinx Dabafluithaas 16 O.s.xunhuaensis Xunhua, China Xunhuafluithaas 17 O. cansus Gansu, China Gansu pika Pika du Gansu Gansu-Pfeifhase Pica de Gansu Gansufluithaas 18 O.c. cansus Gray pika Quillianfluithaas 19 O.c morosa chagrijnig Shensi pika Shaanxifluithaas 20 O.c. stevensi Herbert Stevens Kangding pika Hengduanfluithaas 21 O. nubrica Nubra vallei, India Nubra pika Pika du Nubra Nubra-Pfeifhase Pica de Nubra Nubrafluithaas 22 O.n. nubrica Tuggur pika Ladakhfluithaas 23 O.n. lhasaensis Lhasa, Tibet Lhasa pika Lhasafluithaas 24 O. -
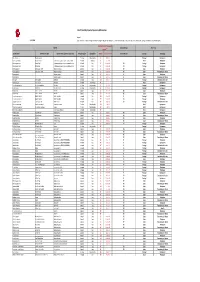
LEPI 2020 Pivot Table VF 20210406 (Version 1).Xlsb
List of Potentially Impacted Species by EDP activities Legend: 31/12/2020 (1) threat levels: i. Critically endangered (CR); ii. Endangered (EN); iii. Vulnerable (VU); iv. Near threatened (NT), Least concern (LC), Data deficient (DD), Not Applicable (NA) and Not Evaluated (NE) IUCN Red List of Threatened Species National Red List Occurrence Species (1) Scientific name Common name (EN) Common name (country of occurrence) Reino/Kingdom Classe/Class 2020‐01 latest assessment National Red List Country Tecnology Dianthus marizii Dianthus marizii ‐ Plantae Magnoliopsida LC 6‐abr‐11 Portugal Hydropower Coendou prehensilis Brazilian Porcupine ouriço‐cacheiro, porco‐espinho, cuandu e cuim Animalia Mammalia LC 10‐jun‐16 Brazil Windpower Hieraetus pennatus Booted Eagle A águia‐pequena, águia‐calçada ou águia‐de‐botas Animalia Aves LC 31‐mar‐15 NT Portugal Windpower Hieraetus pennatus Booted Eagle A águia‐pequena, águia‐calçada ou águia‐de‐botas Animalia Aves LC 31‐mar‐15 NT Portugal Hydropower Merops apiaster European Bee‐eater Abejaruco europeo Animalia Aves LC 31‐mar‐15 NE Spain Windpower Merops apiaster European Bee‐eater Abejaruco europeo Animalia Aves LC 31‐mar‐15 NE Spain Transmision grid‐ Renew Pernis apivorus Abejero europeo Animalia Aves LC 1‐out‐16 LC Spain Windpower Pernis apivorus Abejero europeo Animalia Aves LC 1‐out‐16 LC Spain Transmision grid‐ Renew Otis tarda Great bustard Abetarda Animalia Aves VU 1‐out‐17 EN Portugal Distribution Grid ‐activ Oxydoras niger ripsaw catfish Abotoado Animalia Actinopterygii NE Brazil Hydropower Santolina -

Spain and Portugal, 26 Dec 2019 – 9 Jan 2020. VLADIMIR DINETS
Spain and Portugal, 26 Dec 2019 – 9 Jan 2020. VLADIMIR DINETS Browsing trip reports on mammalwatching.com (my favorite way of self-torture for hardening the spirit), I noticed that there were none from mainland Portugal. So I talked my wife into going there for the winter break; we brought along my daughter and in-laws to make it more entertaining, so all mammalwatching was done at night or at dawn. We spent a few days in western Spain as well. Despite excellent weather I got only 31 species; goodies included Morisco roe deer, Pyrenean desman of the rarely seen western subspecies, Iberian shrew which I missed in 2014 despite much effort, and the recently split Portuguese field vole, the latter two seen within 40 min from each other. We forgot my camera and all flashlights at home, so I have no mammal photos except those taken with my phone. New moon was on December 26. Spain The weather was dry and cold in Spain, with strong temperature inversion in the interior (-2oC at night in Avila but +4 at 1700 m in Sierra de Gredos). Night drives and thermal scoping in fields, pastures and open woodland were remarkably devoid of mammal sightings. Avila: There are small colonies of Mediterranean pine voles in steeper portions of grassy slopes below the city walls, the best one at 40.659619N 4.704030W. Park across the street and watch the slope until you spot a vole; there is street lighting. A thermal scope helps. I saw one at 1 am after 2 hours of watching. -

Spain's Coto Donana & Extremadura
Spain's Coto Donana & Extremadura Naturetrek Tour Report 28 April - 6 May 2018 Booted Eagle Marbled Duck Tour report & images by Simon Tonkin Naturetrek Mingledown Barn Wolf's Lane Chawton Alton Hampshire GU34 3HJ UK T: +44 (0)1962 733051 E: [email protected] W: www.naturetrek.co.uk Tour Report Spain's Coto Donana & Extremadura Tour participants: Simon Tonkin (Leader) with a group of seven Naturetrek clients Day 1 Saturday 28th April Arriving into Sevilla, Simon was on hand to meet the group from their flight and we were soon on the road, although not before getting to grips with Pallid Swifts swerving around the terminal building, providing views even before we got to the minibus. Soon we were out of Sevilla and encountering our first Black Kites and Booted Eagles on route to El Rocìo. As we swung into El Rocìo, two things struck the group - the abundant array of White Stork nests and the swarm of Common House Martins swirling overhead like welcome fireworks! Having checked into our hotel, we were soon out into the sandy streets of El Rocio and birding the freshwater lagoon whilst Simon prepared the picnic of local bread, cheeses and jamon, all washed down with some local red or white wine. Greater Flamingoes and Eurasian Spoonbills were abundant and provided the backdrop as our attention was taken by Great Reed Warblers, Western Swamphens (a recent split from Purple Swamphen), a fantastic pair of Little Bitterns and a single Squacco Heron. Meanwhile the air above was full of hirundines and Black Kites - what a way to begin the tour in pure ornithological elysium! Day 2 Sunday 29th April We set off into the interior of the park with local Doñana guide Rosario who works for Doñana Nature, in whose 4x4 vehicles we can access parts of the park we wouldn’t otherwise see. -

See List of Animal Species of Sierra De Andujar
LIST OF ANIMAL SPECIES OF SIERRA DE ANDUJAR English Latin Español BIRDS AVES Alpine accentor Prunella collaris Acentor alpino Alpine swift Tachimarptis melba Vencejo real Azure-winged magpie Cyanopica cooki Rabilargo Barn owl Tyto alba Lechuza común Bee-eater Merops apiaster Abejaruco común Black kite Milvus migrans Milano negro Black redstart Phoenicurus ochruros Colirrojo tizón Black stork Ciconia nigra Cigüeña negra Black culture Aegypius monachus Buitre negro Black wheatear Oenanthe leucura Collalba negra Blackbird Turdus merula Mirlo común Blackcap Sylvia atricapilla Curruca capirotada Black-eared wheatear Oenanthe hispanica Collalba rubia Blue rock thrush Monticola solitarius Roquero solitario Blue tit Cyanistes caeruleus Herrerillo común Blue-headed wagtail Motacilla flava Lavandera boyera Bonelli´s eagle Aquila fasciata Águila perdicera Bonelli´s warbler Phylloscopus bonelli Mosquitero papialbo Booted eagle Hieraaetus pennatus Águililla calzada Brambling Fringilla montifringilla Pinzón real Bullfinch Pyrrhula pyrrhula Camachuelo común Buzzard Buteo buteo Busardo ratonero Calandra lark Melanocorypha calandra Calandria Cattle egret Bubulcus ibis Garcilla bueyera Cetti´s warbler Cettia cetti Ruiseñor bastardo Chaffinch Fringilla coelebs Pinzón vulgar Chiffchaff Philloscopus collybita Mosquitero común Chough Phyrrhocorax phyrrhocorax Chova piquirroja Cirl bunting Emberiza cirlus Escribano soteño Coal tit Parus ater Carbonero garrapinos Collared turtle dove Streptotelia decaocto Tórtola turca Common sandpiper Actitis hypoleucos Andarríos -

Unmatched DNA Preservation Prove Arctic Hare and Sheep Wool in Norse Greenlandic Textile from “The Farm Beneath the Sand” MARK
Journal of Archaeological Science: Reports 14 (2017) 603–608 Contents lists available at ScienceDirect Journal of Archaeological Science: Reports journal homepage: www.elsevier.com/locate/jasrep Unmatched DNA preservation prove arctic hare and sheep wool in Norse Greenlandic textile from “The Farm Beneath the Sand” MARK Mikkel-Holger S. Sindinga,b,⁎, Filipe Garrett Vieiraa, M. Hayeur Smithc a Centre for GeoGenetics, Natural History Museum of Denmark, Øster Voldgade 5–7, DK-1350 Copenhagen, Denmark b Natural History Museum, University of Oslo, P.O. Box 1172, Blindern, NO-0318 Oslo, Norway c Haffenreffer Museum of Anthropology, Brown University, 300 Tower St Bristol, RI 02809, USA ABSTRACT GUS (Gården under sandet- The Farm Beneath the Sand) is a Greenlandic Norse settlement site 80 km from Nuuk in the former Norse western settlement occupied between 1000 and 1400 CE. Renowned for its excellent pre- servation caused by its interment under large quantities of sand and permafrost after its abandonment, GUS is unique in Norse Greenlandic contexts as perishable materials and artefacts are extremely well preserved. Some aspects of fibre admixtures used in Norse Greenlandic clothing are unknown to us, but of great relevance to understanding the history of the colony and its subsistence practices. Here we present the results of shotgun genomic data from 11 samples originating from ten archaeological textiles from a variety of different Norse Greenlandic sites. The obtained sequences were mapped to mitochondrial genomes of 15 diverse mammals and only samples from GUS had any endogenous DNA (4,5 and 3,5%), resulting in a 70x mt-genome of arctic hare (Lepus arcticus) and a 20x mt-genome of domestic sheep (Ovis aries). -

The Health and Future of the Six Hare Species in Europe: a Closer Look at the Iberian Hare Margarida D
Chapter The Health and Future of the Six Hare Species in Europe: A Closer Look at the Iberian Hare Margarida D. Duarte, Carina L. Carvalho, Fábio Abade dos Santos, Jéssica Monteiro, Madalena Monteiro, Paulo Melo Carvalho, Paula Mendonça, Patrícia Tavares Santos and Pedro C. Melo Abstract Although there are around 40 species of hares in the world divided into three different genera (Lepus, Caprolagus, and Pronolagus), only six species inhabit Europe, all belonging to genus Lepus. The conservation status of these six species was recently revised in the International Conservation Union (IUCN) Red List of Threatened Species. Lepus castroviejoi and L. corsicanus were attributed the status of “vulnerable”. The other four species, L. europaeus, L. timidus, L. capensis, and L. granatensis, were considered of “least concern” although a declining trend was recognized for the last two species’ wild populations. Here we review the major threats to the hare species in Europe, with emphasis on infectious diseases. Furthermore, we present the sanitary data regarding the Iberian hare populations from Portugal, which were severely affected by the emergence of a naturally occur- ring recombinant myxoma virus (MYXV), first reported in mid-2018. The recent detection in 2019 of a leporid herpesvirus (LeHV-5), which pathogenicity appears to be exacerbated in MYXV-infected hares, brings additional concerns to the health and conservation of the Iberian hare. Keywords: hare species, Iberian hare, Lepus granatensis, viral diseases, myxomatosis, myxoma virus, MYXV, rabbit hemorrhagic disease virus, RHDV2, leporid herpesvirus, LeHV-5 1. Introduction 1.1 Geographic distribution in Europe The Lagomorpha order (belonging to the Mammalia class) includes the Ochotonidae family, with one sole genus designated Ochotona, and the Leporidae family, with 11 genera, namely, Pentalagus, Bunolagus, Nesolagus, Romerolagus, Brachylagus, Sylvilagus, Poelagus, Pronolagus, Caprolagus, Oryctolagus, and Lepus. -
Spring in Spain Birding and History Private Tour 2018 Trip Report 19 April - 3 May 2018 (15 Days)
Spring in Spain Birding and History Private Tour 2018 Trip Report 19 April - 3 May 2018 (15 days) Text and pictures by Tour Leader: Yeray Seminario Subalpine Warbler singing its heart away at Monfragüe National Park Spring in Spain - Birding and History Tour 2018 Tour Summary This tour was desiGned for the sprinG season and customized upon a few requirements from the parKcipants, like the inclusion of some historical sites such as La Alhambra and the Roman ruins of Mérida. SprinG is probably the best season to visit Spain, and we focused in all the specialKes of the reGion that would be breedinG at the Kme. Some of the hiGhliGhts of this trip: • The scenery on most of the trip was fantasKc - the copious rains before the trip and the Good weather throuGh most of our iKnerary, made for an unforGeRable journey, with bloominG flowers everywhere. • Monfragüe NP and the plains around Trujillo were very producKve, givinG us good views of the Spanish Imperial Eagle and some distant views, but with excellent behavior, of displayinG male Great Bustard and Lile Bustard. • GenG to see an Iberian Lynx, one of the most criKcally endanGered cats in the world, and the followinG day a Bearded Vulture, was certainly another hiGhliGht. • Fuente de Piedra was parKcularly Good, with a record number of Greater Flamingos breedinG this year, and even Got to see a few Lesser Flamingos. • The Doñana wetlands were teeminG with life, includinG Good numbers of shorebirds and waterfowl, havinG close views of key species like Marbled Teal and White- headed Duck.