Alpha-1 Antitrypsin Inhibits SARS-Cov-2 Infection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Types of Acute Phase Reactants and Their Importance in Vaccination (Review)
BIOMEDICAL REPORTS 12: 143-152, 2020 Types of acute phase reactants and their importance in vaccination (Review) RAFAAT H. KHALIL1 and NABIL AL-HUMADI2 1Department of Biology, College of Science and Technology, Florida Agricultural and Mechanical University, Tallahassee, FL 32307; 2Office of Vaccines, Food and Drug Administration, Center for Biologics Evaluation and Research, Silver Spring, MD 20993, USA Received May 10, 2019; Accepted November 25, 2019 DOI: 10.3892/br.2020.1276 Abstract. Vaccines are considered to be one of the most human and veterinary medicine. Proteins which are expressed cost-effective life-saving interventions in human history. in the acute phase are potential biomarkers for the diagnosis The body's inflammatory response to vaccines has both of inflammatory disease, for example, acute phase proteins desired effects (immune response), undesired effects [(acute (APPs) are indicators of successful organ transplantation phase reactions (APRs)] and trade‑offs. Trade‑offs are and can be used to predict the ameliorative effect of cancer more potent immune responses which may be potentially therapy (1,2). APPs are primarily synthesized in hepatocytes. difficult to separate from potent acute phase reactions. The acute phase response is a spontaneous reaction triggered Thus, studying acute phase proteins (APPs) during vaccina- by disrupted homeostasis resulting from environmental distur- tion may aid our understanding of APRs and homeostatic bances (3). Acute phase reactions (APRs) usually stabilize changes which can result from inflammatory responses. quickly, after recovering from a disruption to homeostasis Depending on the severity of the response in humans, these within a few days to weeks; however, APPs expression levels reactions can be classified as major, moderate or minor. -

Lactoferrin and Its Detection Methods: a Review
nutrients Review Lactoferrin and Its Detection Methods: A Review Yingqi Zhang, Chao Lu and Jin Zhang * Department of Chemical and Biochemical Engineering, University of Western Ontario, London, ON N6A 5B9, Canada; [email protected] (Y.Z.); [email protected] (C.L.) * Correspondence: [email protected] Abstract: Lactoferrin (LF) is one of the major functional proteins in maintaining human health due to its antioxidant, antibacterial, antiviral, and anti-inflammatory activities. Abnormal levels of LF in the human body are related to some serious diseases, such as inflammatory bowel disease, Alzheimer’s disease and dry eye disease. Recent studies indicate that LF can be used as a biomarker for diagnosis of these diseases. Many methods have been developed to detect the level of LF. In this review, the biofunctions of LF and its potential to work as a biomarker are introduced. In addition, the current methods of detecting lactoferrin have been presented and discussed. We hope that this review will inspire efforts in the development of new sensing systems for LF detection. Keywords: lactoferrin; biomarkers; immunoassay; instrumental analysis; sensor 1. Introduction Lactoferrin (known as lactotransferrin, LF), with a molecular weight of about 80 kDa, is a functional glycoprotein, which contains about 690 amino acid residues. It was first isolated from bovine milk by Sorensen in 1939 and was first isolated from human milk by Citation: Zhang, Y.; Lu, C.; Zhang, J. Johanson in 1960 [1,2]. The three-dimensional structure of LF has been unveiled by high Lactoferrin and Its Detection resolution X-ray crystallographic analysis, and it consists of two homologous globular lobes Methods: A Review. -

Influence of Infection and Inflammation on Biomarkers of Nutritional Status
A2.4 INFLUENCE OF INFECTION AND INFLAMMATION ON BIOMARKERS OF NUTRITIONAL STATUS A2.4 Influence of infection and inflammation on biomarkers of nutritional status with an emphasis on vitamin A and iron David I. Thurnham1 and George P. McCabe2 1 Northern Ireland Centre for Food and Health, University of Ulster, Coleraine, United Kingdom of Great Britain and Northern Ireland 2 Statistics Department, Purdue University, West Lafayette, Indiana, United States of America Corresponding author: David I. Thurnham; [email protected] Suggested citation: Thurnham DI, McCabe GP. Influence of infection and inflammation on biomarkers of nutritional status with an emphasis on vitamin A and iron. In: World Health Organization. Report: Priorities in the assessment of vitamin A and iron status in populations, Panama City, Panama, 15–17 September 2010. Geneva, World Health Organization, 2012. Abstract n Many plasma nutrients are influenced by infection or tissue damage. These effects may be passive and the result of changes in blood volume and capillary permeability. They may also be the direct effect of metabolic alterations that depress or increase the concentration of a nutrient or metabolite in the plasma. Where the nutrient or metabolite is a nutritional biomarker as in the case of plasma retinol, a depression in retinol concentrations will result in an overestimate of vitamin A deficiency. In contrast, where the biomarker is increased due to infection as in the case of plasma ferritin concentrations, inflammation will result in an underestimate of iron deficiency. Infection and tissue damage can be recognized by their clinical effects on the body but, unfortunately, subclinical infection or inflammation can only be recognized by measur- ing inflammation biomarkers in the blood. -

The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria
Downloaded from orbit.dtu.dk on: Oct 02, 2021 The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria Kell, Douglas B.; Heyden, Eugene L.; Pretorius, Etheresia Published in: Frontiers in Immunology Link to article, DOI: 10.3389/fimmu.2020.01221 Publication date: 2020 Document Version Publisher's PDF, also known as Version of record Link back to DTU Orbit Citation (APA): Kell, D. B., Heyden, E. L., & Pretorius, E. (2020). The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Frontiers in Immunology, 11, [1221]. https://doi.org/10.3389/fimmu.2020.01221 General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. Users may download and print one copy of any publication from the public portal for the purpose of private study or research. You may not further distribute the material or use it for any profit-making activity or commercial gain You may freely distribute the URL identifying the publication in the public portal If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. HYPOTHESIS AND THEORY published: 28 May 2020 doi: 10.3389/fimmu.2020.01221 The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria Douglas B. -

Alpha -Antitrypsin Deficiency
The new england journal of medicine Review Article Dan L. Longo, M.D., Editor Alpha1-Antitrypsin Deficiency Pavel Strnad, M.D., Noel G. McElvaney, D.Sc., and David A. Lomas, Sc.D. lpha1-antitrypsin (AAT) deficiency is one of the most common From the Department of Internal Med genetic diseases. Most persons carry two copies of the wild-type M allele icine III, University Hospital RWTH of SERPINA1, which encodes AAT, and have normal circulating levels of the (Rheinisch–Westfälisch Technische Hoch A schule) Aachen, Aachen, Germany (P.S.); protein. Ninety-five percent of severe cases of AAT deficiency result from the homo- the Irish Centre for Genetic Lung Dis zygous substitution of a single amino acid, Glu342Lys (the Z allele), which is present ease, Royal College of Surgeons in Ire in 1 in 25 persons of European descent (1 in 2000 persons of European descent land, Beaumont Hospital, Dublin (N.G.M.); and UCL Respiratory, Division of Medi are homozygotes). Mild AAT deficiency typically results from a different amino cine, Rayne Institute, University College acid replacement, Glu264Val (the S allele), which is found in 1 in 4 persons in the London, London (D.A.L.). Address re Iberian peninsula. However, many other alleles have been described that have vari- print requests to Dr. Lomas at UCL Re spiratory, Rayne Institute, University Col able effects, such as a lack of protein production (null alleles), production of mis- lege London, London WC1E 6JF, United folded protein, or no effect on the level or function of circulating AAT (Table 1). Kingdom, or at d . -

Identification of Key Pathways and Genes in Dementia Via Integrated Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of Key Pathways and Genes in Dementia via Integrated Bioinformatics Analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract To provide a better understanding of dementia at the molecular level, this study aimed to identify the genes and key pathways associated with dementia by using integrated bioinformatics analysis. Based on the expression profiling by high throughput sequencing dataset GSE153960 derived from the Gene Expression Omnibus (GEO), the differentially expressed genes (DEGs) between patients with dementia and healthy controls were identified. With DEGs, we performed a series of functional enrichment analyses. Then, a protein–protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network was constructed, analyzed and visualized, with which the hub genes miRNAs and TFs nodes were screened out. Finally, validation of hub genes was performed by using receiver operating characteristic curve (ROC) analysis. -
2019 Laboratory Report
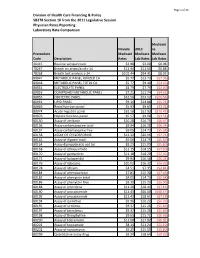
Page 1 of 24 Division of Health Care Financing & Policy SB278 Section 16 from the 2011 Legislative Session Physician Rates Reporting Laboratory Rate Comparison Medicaid Nevada 2019 vs. Proccedure Medicaid Medicare Medicare Code Description Rates Lab Rates Lab Rates 36415 Routine venipuncture $3.98 $3.00 $0.98 78267 Breath tst attain/anal c-14 $11.94 $11.06 $0.88 78268 Breath test analysis c-14 $102.44 $94.41 $8.03 80047 METABOLIC PANEL IONIZED CA $5.77 $13.73 ($7.96) 80048 METABOLIC PANEL TOTAL CA $5.77 $9.40 ($3.63) 80051 ELECTROLYTE PANEL $4.79 $7.79 ($3.00) 80053 COMPREHEN METABOLIC PANEL $7.21 $11.74 ($4.53) 80055 OBSTETRIC PANEL $32.56 $53.12 ($20.56) 80061 LIPID PANEL $9.14 $14.88 ($5.74) 80069 Renal function panel $5.93 $9.65 ($3.72) 80074 Acute hepatitis panel $32.50 $52.93 ($20.43) 80076 Hepatic function panel $5.57 $9.08 ($3.51) 80150 Assay of amikacin $10.28 $16.75 ($6.47) 80156 Assay carbamazepine total $9.94 $16.18 ($6.24) 80157 Assay carbamazepine free $9.05 $14.73 ($5.68) 80158 ASSAY OF CYCLOSPORINE $12.32 $20.06 ($7.74) 80162 Assay of digoxin total $9.06 $14.75 ($5.69) 80164 Assay dipropylacetic acd tot $9.25 $15.05 ($5.80) 80168 Assay of ethosuximide $11.15 $18.15 ($7.00) 80170 Assay of gentamicin $11.18 $18.20 ($7.02) 80173 Assay of haloperidol $9.94 $16.18 ($6.24) 80176 Assay of lidocaine $10.02 $16.32 ($6.30) 80178 Assay of lithium $4.51 $7.35 ($2.84) 80184 Assay of phenobarbital $7.81 $15.30 ($7.49) 80185 Assay of phenytoin total $9.05 $14.73 ($5.68) 80186 Assay of phenytoin free $9.39 $15.29 ($5.90) 80188 Assay of -

Review Article C-Lobe of Lactoferrin: the Whole Story of the Half-Molecule
Hindawi Publishing Corporation Biochemistry Research International Volume 2013, Article ID 271641, 8 pages http://dx.doi.org/10.1155/2013/271641 Review Article C-Lobe of Lactoferrin: The Whole Story of the Half-Molecule Sujata Sharma, Mau Sinha, Sanket Kaushik, Punit Kaur, and Tej P. Singh DepartmentofBiophysics,AllIndiaInstituteofMedicalSciences,NewDelhi110029,India Correspondence should be addressed to Tej P. Singh; [email protected] Received 24 January 2013; Accepted 21 March 2013 Academic Editor: Andrei Surguchov Copyright © 2013 Sujata Sharma et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Lactoferrin is an iron-binding diferric glycoprotein present in most of the exocrine secretions. The major role of lactoferrin, which is found abundantly in colostrum, is antimicrobial action for the defense of mammary gland and the neonates. Lactoferrin consists of two equal halves, designated as N-lobe and C-lobe, each of which contains one iron-binding site. While the N-lobe of lactoferrin has been extensively studied and is known for its enhanced antimicrobial effect, the C-lobe of lactoferrin mediates various therapeutic functions which are still being discovered. The potential of the C-lobe in the treatment of gastropathy, diabetes, and corneal wounds and injuries has been indicated. This review provides the details of the proteolytic preparation of C-lobe, and interspecies comparisons of its sequence and structure, as well as the scope of its therapeutic applications. 1. Lactoferrin: A Bilobal Protein edge on the structures of two independent lobes upon cleav- age of lactoferrin has been scarce. -

Haptoglobin-Matrix Metalloproteinase 9 Complex As a Biomarker for Acute Inflammation in Cattle
Haptoglobin-Matrix Metalloproteinase 9 Complex as a Biomarker for Acute Inflammation in Cattle THESIS Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By Charles Austin Hinds D.V.M Graduate Program in Veterinary Clinical Sciences The Ohio State University 2011 Master's Examination Committee: Jeffrey Lakritz, Advisor Andrew Niehaus Christopher Premanandan Paivi Rajala-Schultz D. Michael Rings Copyrighted by Charles Austin Hinds 2011 Abstract Bovine respiratory disease (BRD) is a major cause of economic loss in feedlots in the United Sates. These losses are associated not only with morbidity and mortality, but also the expense of using antimicrobial drugs unnecessarily. One of the recurring problems is the inability to diagnose and therefore treat respiratory disease appropriately. An Hp-MMP 9 protein complex has been identified in neutrophil granules and in the serum of cattle with acute bacterial sepsis. The purpose of this project was to evaluate the utility of an Hp-MMP 9 complex ELISA in the diagnosis of acute septic inflammation in cattle. Three experiments were performed. The first experiment was designed to determine whether Hp-MMP 9 could be used in the prediction of BRD in calves recently admitted to feedlots. Using health, treatment and weight gain data, our aim was to determine whether Hp-MMP 9 could predict which calves would be identified with clinical respiratory disease and would require therapy in the days following sample collection. We compared serum concentrations of Hp to Hp-MMP 9 to assess how well the complex performed in these animals. -

Lactoferrin (Mouse) ELISA Kit Rev09/19
FOR RESEARCH USE ONLY! Lactoferrin (Mouse) ELISA Kit rev09/19 (Catalog # E4727-100; 96 assays; Storage at 4ºC) I. Introduction: Lactoferrin is a glycoprotein present in exocrine secretions and in the secondary granules of polymorphonuclear neutrophils. It is the iron-binding protein in milk. Lactoferrin is known to regulate intestinal iron absorption, and to participate in the defense against bacteria. BioVision’s Lactoferrin ELISA kit uses the Sandwich-ELISA principle. The micro ELISA plate provided in the kit has been pre-coated with an antibody specific to mouse Lactoferrin. On addition of standards or samples to the micro ELISA plate, they react with Biotinylated detection antibody specific for mouse Lactoferrin and Avidin-Horseradish Peroxidase (HRP) conjugate, which on the addition of substrate gives blue color. The enzyme-substrate reaction is terminated by the addition of stop solution to turn the reaction yellow. The optical density (OD) is measured spectrophotometrically at a wavelength of 450 nm. The OD value is proportional to the concentration of mouse LTF/LF. The concentration of mouse Lactoferrin in the samples can be calculated by comparing the OD of the samples to the standard curve. II. Applications: • in vitro quantitative determination of Mouse LTF/LF concentrations in serum, plasma and other biological fluids. • Sensitivity: 37.50 pg/ml • Coefficient of variation is < 10% • Detection Range: 62.50 - 4000 pg/ml • Specificity: This kit recognizes Mouse LTF/LF in samples. No significant cross-reactivity or interference -

Blood Serum Acute Phase Proteins and Iron Dynamics During Acute Phase Response of Salmonella Enterica Serotype Dublin Experiment
Veterinary Immunology and Immunopathology 203 (2018) 30–39 Contents lists available at ScienceDirect Veterinary Immunology and Immunopathology journal homepage: www.elsevier.com/locate/vetimm Blood serum acute phase proteins and iron dynamics during acute phase response of Salmonella enterica serotype Dublin experimentally infected T buffalo calves ⁎ André M. Santanaa, , Daniela G. Silvaa, Funmilola C. Thomasb, Priscila A. Bernardesa, Lucas J.L. Pizauroa, Clarissa H. Santanaa, Richard J.S. Burchmorec, Peter D. Eckersalld, José J. Fagliaria a Department of Veterinary Clinic and Surgery, School of Agricultural and Veterinary Sciences, São Paulo State University (FCAV/UNESP), Jaboticabal, SP, Brazil b Department of Veterinary Physiology and Pharmacology, College of Veterinary Medicine, Federal University of Agriculture, Abeokuta, Nigeria c Institute of Infection, Immunity and Inflammation, Glasgow Polyomics Facility, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom d Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom ARTICLE INFO ABSTRACT Keywords: The study aimed to evaluate clinical signs, blood serum acute phase proteins (APP) and iron dynamics during the Ceruloplasmin acute phase response (APR) of Salmonella Dublin experimentally infected Murrah buffalo calves. Six buffalo Electrophoresis calves constituted the control group (CNT) and six were orally inoculate with 108 CFU of S. Dublin (INF). Clinical Fibrinogen evaluation was performed, rectal swabs to detect S. Dublin strains were collected and venous blood was sampled Haptoglobin before and throughout seven days after inoculation. The APP fractions β-haptoglobin, α-haptoglobin, cer- Transferrin uloplasmin and transferrin were analyzed by 1-D and 2-D electrophoresis. -

Alpha-1 Antitrypsin Therapy Modulates the Neutrophil Membrane Proteome and Secretome
Early View Original article Alpha-1 antitrypsin therapy modulates the neutrophil membrane proteome and secretome Mark P. Murphy, Thomas McEnery, Karen McQuillan, Oisín F. McElvaney, Oliver J. McElvaney, Sarah Landers, Orla Coleman, Anchalin Bussayajirapong, Padraig Hawkins, Michael Henry, Paula Meleady, Emer P. Reeves, Noel G. McElvaney Please cite this article as: Murphy MP, McEnery T, McQuillan K, et al. Alpha-1 antitrypsin therapy modulates the neutrophil membrane proteome and secretome. Eur Respir J 2020; in press (https://doi.org/10.1183/13993003.01678-2019). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2020 Alpha-1 antitrypsin therapy modulates the neutrophil membrane proteome and secretome Mark P. Murphy,1 Thomas McEnery,1 Karen McQuillan,1 Oisín F. McElvaney,1 Oliver J. McElvaney,1 Sarah Landers,1 Orla Coleman,2 Anchalin Bussayajirapong,1 Padraig Hawkins,1 Michael Henry,2 Paula Meleady,2 Emer P. Reeves,1 Noel G. McElvaney1 1 Irish Centre for Genetic Lung Disease, Department of Medicine, Royal College of Surgeons in Ireland, Education and Research Centre, Beaumont Hospital, Dublin 9, Ireland. 2 National Institute for Cellular Biotechnology, Dublin City University, Glasnevin, Dublin 9, Ireland. Correspondence should be addressed to: Dr Emer P. Reeves, Ph.D., Irish Centre for Genetic Lung Disease, Royal College of Surgeons in Ireland, Education and Research Centre, Beaumont Hospital, Dublin 9, Ireland.