Phylogenetic Comparison and Splice Site Conservation of Eukaryotic U1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Unrip (STRAP) (NM 007178) Human Tagged ORF Clone Lentiviral Particle Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC209149L3V Unrip (STRAP) (NM_007178) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: Unrip (STRAP) (NM_007178) Human Tagged ORF Clone Lentiviral Particle Symbol: STRAP Synonyms: MAWD; PT-WD; UNRIP Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) ACCN: NM_007178 ORF Size: 1050 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC209149). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_007178.2 RefSeq Size: 1924 bp RefSeq ORF: 1053 bp Locus ID: 11171 UniProt ID: Q9Y3F4 Domains: WD40 Protein Families: Druggable Genome MW: 38.4 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 Unrip (STRAP) (NM_007178) Human Tagged ORF Clone Lentiviral Particle – RC209149L3V Gene Summary: The SMN complex plays a catalyst role in the assembly of small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Immune Adaptation to Chronic Intense Exercise Training: New Microarray Evidence Dongmei Liu1†, Ru Wang1†, Ana R
Liu et al. BMC Genomics (2017) 18:29 DOI 10.1186/s12864-016-3388-5 RESEARCHARTICLE Open Access Immune adaptation to chronic intense exercise training: new microarray evidence Dongmei Liu1†, Ru Wang1†, Ana R. Grant2, Jinming Zhang3, Paul M. Gordon4, Yuqin Wei1 and Peijie Chen1* Abstract Background: Endurance exercise training, especially the high-intensity training, exhibits a strong influence on the immune system. However, the mechanisms underpinning the immune-regulatory effect of exercise remain unclear. Consequently, we chose to investigate the alterations in the transcriptional profile of blood leukocytes in young endurance athletes as compared with healthy sedentary controls, using Affymetrix human gene 1.1 ST array. Results: Group differences in the transcriptome were analyzed using Intensity-based Hierarchical Bayes method followed by a Logistic Regression-based gene set enrichment method. We identified 72 significant transcripts differentially expressed in the leukocyte transcriptome of young endurance athletes as compared with non-athlete controls with a false discovery rate (FDR) < 0.05, comprising mainly the genes encoding ribosomal proteins and the genes involved in mitochondrial oxidative phosphorylation. Gene set enrichment analysis identified three major gene set clusters: two were up-regulated in athletes including gene translation and ribosomal protein production, and mitochondria oxidative phosphorylation and biogenesis; one gene set cluster identified as transcriptionally downregulated in athletes was related to inflammation and immune activity. Conclusion: Our data indicates that in young healthy individuals, intense endurance exercise training (exemplifed by athletic training) can chronically induce transcriptional changes in the peripheral blood leukocytes, upregulating genes related to protein production and mitochondrial energetics, and downregulating genes involved in inflammatory response. -

U1 Snrnp Regulates Chromatin Retention of Noncoding Rnas
bioRxiv preprint doi: https://doi.org/10.1101/310433; this version posted April 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. U1 snRNP regulates chromatin retention of noncoding RNAs Yafei Yin,1,* J. Yuyang Lu,1 Xuechun Zhang,1 Wen Shao,1 Yanhui Xu,1 Pan Li,1,2 Yantao Hong,1 Qiangfeng Cliff Zhang,2 and Xiaohua Shen 1,3,* 1 Tsinghua-Peking Center for Life Sciences, School of Medicine and School of Life Sciences, Tsinghua University, Beijing 100084, China 2 MOE Key Laboratory of Bioinformatics, Beijing Advanced Innovation Center for Structural Biology, Center for Synthetic and Systems Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China 3 Lead Contact *Correspondence: [email protected] (Y.Y.), [email protected] (X.S.) Running title: U1 snRNP regulates ncRNA-chromatin retention Keywords: RNA-chromatin localization, U1 snRNA, lncRNA, PROMPTs and eRNA, splicing 1 bioRxiv preprint doi: https://doi.org/10.1101/310433; this version posted April 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Thousands of noncoding transcripts exist in mammalian genomes, and they preferentially localize to chromatin. Here, to identify cis-regulatory elements that control RNA-chromatin association, we developed a high-throughput method named RNA element for subcellular localization by sequencing (REL-seq). Coupling REL-seq with random mutagenesis (mutREL-seq), we discovered a key 7-nt U1 recognition motif in chromatin-enriched RNA elements. -

U1 Snrnp Regulates Chromatin Retention of Noncoding Rnas
Article U1 snRNP regulates chromatin retention of noncoding RNAs https://doi.org/10.1038/s41586-020-2105-3 Yafei Yin1 ✉, J. Yuyang Lu1, Xuechun Zhang1, Wen Shao1, Yanhui Xu1, Pan Li1,2, Yantao Hong1, Li Cui1, Ge Shan3, Bin Tian4, Qiangfeng Cliff Zhang1,2 & Xiaohua Shen1 ✉ Received: 26 November 2018 Accepted: 9 January 2020 Long noncoding RNAs (lncRNAs) and promoter- or enhancer-associated unstable Published online: xx xx xxxx transcripts locate preferentially to chromatin, where some regulate chromatin structure, Check for updates transcription and RNA processing1–13. Although several RNA sequences responsible for nuclear localization have been identifed—such as repeats in the lncRNA Xist and Alu-like elements in long RNAs14–16—how lncRNAs as a class are enriched at chromatin remains unknown. Here we describe a random, mutagenesis-coupled, high-throughput method that we name ‘RNA elements for subcellular localization by sequencing’ (mutREL-seq). Using this method, we discovered an RNA motif that recognizes the U1 small nuclear ribonucleoprotein (snRNP) and is essential for the localization of reporter RNAs to chromatin. Across the genome, chromatin-bound lncRNAs are enriched with 5′ splice sites and depleted of 3′ splice sites, and exhibit high levels of U1 snRNA binding compared with cytoplasm-localized messenger RNAs. Acute depletion of U1 snRNA or of the U1 snRNP protein component SNRNP70 markedly reduces the chromatin association of hundreds of lncRNAs and unstable transcripts, without altering the overall transcription rate in cells. In addition, rapid degradation of SNRNP70 reduces the localization of both nascent and polyadenylated lncRNA transcripts to chromatin, and disrupts the nuclear and genome-wide localization of the lncRNA Malat1. -

Anti-SNRPD3 Antibody (ARG55763)
Product datasheet [email protected] ARG55763 Package: 100 μl anti-SNRPD3 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes SNRPD3 Tested Reactivity Hu Predict Reactivity Ms, Xenopus Tested Application FACS, IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name SNRPD3 Antigen Species Human Immunogen KLH-conjugated synthetic peptide corresponding to aa. 99-126 (C-terminus) of Human SNRPD3. Conjugation Un-conjugated Alternate Names Small nuclear ribonucleoprotein Sm D3; Sm-D3; snRNP core protein D3; SMD3 Application Instructions Application table Application Dilution FACS 1:10 - 1:50 IHC-P 1:50 - 1:100 WB 1:1000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control MCF7 Calculated Mw 14 kDa Properties Form Liquid Purification Purification with Protein A and immunogen peptide. Buffer PBS and 0.09% (W/V) Sodium azide. Preservative 0.09% (W/V) Sodium azide. Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/3 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Database links GeneID: 6634 Human Swiss-port # P62318 Human Gene Symbol SNRPD3 Gene Full Name small nuclear ribonucleoprotein D3 polypeptide 18kDa Background This gene encodes a core component of the spliceosome, which is a nuclear ribonucleoprotein complex that functions in pre-mRNA splicing. -

Snrpg (NM 026506) Mouse Tagged ORF Clone – MR220563 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for MR220563 Snrpg (NM_026506) Mouse Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: Snrpg (NM_026506) Mouse Tagged ORF Clone Tag: Myc-DDK Symbol: Snrpg Synonyms: 2810024K17Rik; AL022803; sm-G; SMG; snRNP-G Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >MR220563 representing NM_026506 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGAGCAAAGCCCACCCTCCCGAGCTGAAGAAGTTTATGGACAAGAAGTTATCATTGAAGTTAAACGGTG GCAGGCATGTCCAAGGAATACTGCGGGGCTTTGATCCCTTTATGAACCTTGTGATTGACGAGTGTGTGGA GATGGCAACCAGTGGGCAACAGAACAACATTGGCATGGTGGTCATCCGAGGAAACAGCATCATCATGTTA GAAGCCTTGGAAAGAGTC ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA Protein Sequence: >MR220563 representing NM_026506 Red=Cloning site Green=Tags(s) MSKAHPPELKKFMDKKLSLKLNGGRHVQGILRGFDPFMNLVIDECVEMATSGQQNNIGMVVIRGNSIIML EALERV TRTRPLEQKLISEEDLAANDILDYKDDDDKV Chromatograms: https://cdn.origene.com/chromatograms/mm9051_f09.zip Restriction Sites: SgfI-MluI This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 Snrpg (NM_026506) Mouse -

Large-Scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks
Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén1,*, Babita Singh1,*, Belén Miñana1,2, Amadís Pagès1, Francesca Mateo3, Miguel Angel Pujana3, Juan Valcárcel1,2,4, Eduardo Eyras1,4,5 1Universitat Pompeu Fabra, Dr. Aiguader 88, E08003 Barcelona, Spain 2Centre for Genomic Regulation, Dr. Aiguader 88, E08003 Barcelona, Spain 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain. 4Catalan Institution for Research and Advanced Studies, Passeig Lluís Companys 23, E08010 Barcelona, Spain *Equal contribution 5Correspondence to: [email protected] Keywords: alternative splicing, RNA binding proteins, splicing networks, cancer 1 Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. We systematically analyzed mutation, copy number and gene expression patterns of 1348 RNA-binding protein (RBP) genes in 11 solid tumor types, together with alternative splicing changes in these tumors and the enrichment of binding motifs in the alternatively spliced sequences. Our comprehensive study reveals widespread alterations in the expression of RBP genes, as well as novel mutations and copy number variations in association with multiple alternative splicing changes in cancer drivers and oncogenic pathways. Remarkably, the altered splicing patterns in several tumor types recapitulate those of undifferentiated cells. -

Downregulation of SNRPG Induces Cell Cycle Arrest and Sensitizes Human Glioblastoma Cells to Temozolomide by Targeting Myc Through a P53-Dependent Signaling Pathway
Cancer Biol Med 2020. doi: 10.20892/j.issn.2095-3941.2019.0164 ORIGINAL ARTICLE Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway Yulong Lan1,2*, Jiacheng Lou2*, Jiliang Hu1, Zhikuan Yu1, Wen Lyu1, Bo Zhang1,2 1Department of Neurosurgery, Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen 518020, China;2 Department of Neurosurgery, The Second Affiliated Hospital of Dalian Medical University, Dalian 116023, China ABSTRACT Objective: Temozolomide (TMZ) is commonly used for glioblastoma multiforme (GBM) chemotherapy. However, drug resistance limits its therapeutic effect in GBM treatment. RNA-binding proteins (RBPs) have vital roles in posttranscriptional events. While disturbance of RBP-RNA network activity is potentially associated with cancer development, the precise mechanisms are not fully known. The SNRPG gene, encoding small nuclear ribonucleoprotein polypeptide G, was recently found to be related to cancer incidence, but its exact function has yet to be elucidated. Methods: SNRPG knockdown was achieved via short hairpin RNAs. Gene expression profiling and Western blot analyses were used to identify potential glioma cell growth signaling pathways affected by SNRPG. Xenograft tumors were examined to determine the carcinogenic effects of SNRPG on glioma tissues. Results: The SNRPG-mediated inhibitory effect on glioma cells might be due to the targeted prevention of Myc and p53. In addition, the effects of SNRPG loss on p53 levels and cell cycle progression were found to be Myc-dependent. Furthermore, SNRPG was increased in TMZ-resistant GBM cells, and downregulation of SNRPG potentially sensitized resistant cells to TMZ, suggesting that SNRPG deficiency decreases the chemoresistance of GBM cells to TMZ via the p53 signaling pathway. -

Downloaded from the TCGA Data Portal Website ( Cell Proliferation Was Determined Using an MTT Assay
ORIGINAL RESEARCH published: 28 August 2020 doi: 10.3389/fonc.2020.01468 Clinical Significance, Cellular Function, and Potential Molecular Pathways of CCT7 in Endometrial Cancer Liwen Wang 1,2†, Wei Zhou 1†, Hui Li 2,3, Hui Yang 1 and Nianchun Shan 1* 1 Department of Gynecology and Obstetrics, Xiangya Hospital, Central South University, Changsha, China, 2 Xiangya School of Medicine, Central South University, Changsha, China, 3 Department of Reproductive, Xiangya Hospital, Central South University, Changsha, China Objective: Endometrial cancer (EC) is a common gynecologic malignancy; myometrial Edited by: invasion (MI) is a typical approach of EC spreads and an important index to assess tumor Sarah M. Temkin, metastasis and outcome in EC patients. CCT7 is a member of the TCP1 chaperone Virginia Commonwealth University, family, involved in cytoskeletal protein folding and unfolding. In this study, the role of United States CCT7 in EC development was investigated. Reviewed by: Syed S. Islam, Methods: Clinical data for 87 EC cases and expression of CCT7 were analyzed. CCT7 King Faisal Specialist Hospital & Research Centre, Saudi Arabia was knocked out using siRNA-CCT7 in Ishikawa and RL95-2 cells, and their function Elena Gershtein, about proliferation, apoptosis, and invasion was further tested. Bioinformatics methods Russian Cancer Research Center NN were used to predict the potential pathways of CCT7 in EC development. Blokhin, Russia *Correspondence: Results: The rates of CCT7-positive cells in EC and adjacent normal endometrium Nianchun Shan tissues had a significant difference (67.8 vs. 51.4%, p = 0.035), and the expression [email protected] rate increased from low to high pathological stage (39.7% in the I/II stage, 71.4% in the †These authors have contributed III/IV stage, p = 0.029). -

Ribosome Profiling Reveals an Important Role for Translational Control in Circadian Gene Expression
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Research Ribosome profiling reveals an important role for translational control in circadian gene expression Christopher Jang,1,4 Nicholas F. Lahens,2,4 John B. Hogenesch,2 and Amita Sehgal1,3 1Department of Neuroscience, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania 19104, USA; 2Department of Systems Pharmacology and Translational Therapeutics, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania 19104, USA; 3Howard Hughes Medical Institute, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania 19104, USA Physiological and behavioral circadian rhythms are driven by a conserved transcriptional/translational negative feedback loop in mammals. Although most core clock factors are transcription factors, post-transcriptional control introduces delays that are critical for circadian oscillations. Little work has been done on circadian regulation of translation, so to address this deficit we conducted ribosome profiling experiments in a human cell model for an autonomous clock. We found that most rhythmic gene expression occurs with little delay between transcription and translation, suggesting that the lag in the ac- cumulation of some clock proteins relative to their mRNAs does not arise from regulated translation. Nevertheless, we found that translation occurs in a circadian fashion for many genes, sometimes imposing an additional level of control on rhythmically expressed mRNAs and, in other cases, conferring rhythms on noncycling mRNAs. Most cyclically tran- scribed RNAs are translated at one of two major times in a 24-h day, while rhythmic translation of most noncyclic RNAs is phased to a single time of day. -
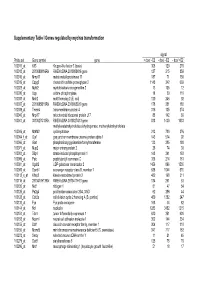
Supplementary Table I Genes Regulated by Myc/Ras Transformation
Supplementary Table I Genes regulated by myc/ras transformation signal Probe set Gene symbol gene + dox/ - E2 - dox/ - E2 - dox/ +E2 100011_at Klf3 Kruppel-like factor 3 (basic) 308 120 270 100013_at 2010008K16Rik RIKEN cDNA 2010008K16 gene 127 215 859 100016_at Mmp11 matrix metalloproteinase 11 187 71 150 100019_at Cspg2 chondroitin sulfate proteoglycan 2 1148 342 669 100023_at Mybl2 myeloblastosis oncogene-like 2 13 105 12 100030_at Upp uridine phosphorylase 18 39 110 100033_at Msh2 mutS homolog 2 (E. coli) 120 246 92 100037_at 2310005B10Rik RIKEN cDNA 2310005B10 gene 178 351 188 100039_at Tmem4 transmembrane protein 4 316 135 274 100040_at Mrpl17 mitochondrial ribosomal protein L17 88 142 89 100041_at 3010027G13Rik RIKEN cDNA 3010027G13 gene 818 1430 1033 methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate 100046_at Mthfd2 cyclohydrolase 213 720 376 100064_f_at Gja1 gap junction membrane channel protein alpha 1 143 574 92 100066_at Gart phosphoribosylglycinamide formyltransferase 133 385 180 100071_at Mup2 major urinary protein 2 26 74 30 100081_at Stip1 stress-induced phosphoprotein 1 148 341 163 100089_at Ppic peptidylprolyl isomerase C 358 214 181 100091_at Ugalt2 UDP-galactose translocator 2 1456 896 1530 100095_at Scarb1 scavenger receptor class B, member 1 638 1044 570 100113_s_at Kifap3 kinesin-associated protein 3 482 168 311 100116_at 2810417H13Rik RIKEN cDNA 2810417H13 gene 134 261 53 100120_at Nid1 nidogen 1 81 47 54 100125_at Pa2g4 proliferation-associated 2G4, 38kD 62 286 44 100128_at Cdc2a cell division cycle 2 homolog A (S. pombe) 459 1382 247 100133_at Fyn Fyn proto-oncogene 168 44 82 100144_at Ncl nucleolin 1283 3452 1215 100151_at Tde1 tumor differentially expressed 1 620 351 620 100153_at Ncam1 neural cell adhesion molecule 1 302 144 234 100155_at Ddr1 discoidin domain receptor family, member 1 304 117 310 100156_at Mcmd5 mini chromosome maintenance deficient 5 (S.