Genetic Characterization of Hantaviruses Isolated from Rodents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Educated Youth Should Go to the Rural Areas: a Tale of Education, Employment and Social Values*
Educated Youth Should Go to the Rural Areas: A Tale of Education, Employment and Social Values* Yang You† Harvard University This draft: July 2018 Abstract I use a quasi-random urban-dweller allocation in rural areas during Mao’s Mass Rustication Movement to identify human capital externalities in education, employment, and social values. First, rural residents acquired an additional 0.1-0.2 years of education from a 1% increase in the density of sent-down youth measured by the number of sent-down youth in 1969 over the population size in 1982. Second, as economic outcomes, people educated during the rustication period suffered from less non-agricultural employment in 1990. Conversely, in 2000, they enjoyed increased hiring in all non-agricultural occupations and lower unemployment. Third, sent-down youth changed the social values of rural residents who reported higher levels of trust, enhanced subjective well-being, altered trust from traditional Chinese medicine to Western medicine, and shifted job attitudes from objective cognitive assessments to affective job satisfaction. To explore the mechanism, I document that sent-down youth served as rural teachers with two new county-level datasets. Keywords: Human Capital Externality, Sent-down Youth, Rural Educational Development, Employment Dynamics, Social Values, Culture JEL: A13, N95, O15, I31, I25, I26 * This paper was previously titled and circulated, “Does living near urban dwellers make you smarter” in 2017 and “The golden era of Chinese rural education: evidence from Mao’s Mass Rustication Movement 1968-1980” in 2015. I am grateful to Richard Freeman, Edward Glaeser, Claudia Goldin, Wei Huang, Lawrence Katz, Lingsheng Meng, Nathan Nunn, Min Ouyang, Andrei Shleifer, and participants at the Harvard Economic History Lunch Seminar, Harvard Development Economics Lunch Seminar, and Harvard China Economy Seminar, for their helpful comments. -

Changchun–Harbin Expressway Project
Performance Evaluation Report Project Number: PPE : PRC 30389 Loan Numbers: 1641/1642 December 2006 People’s Republic of China: Changchun–Harbin Expressway Project Operations Evaluation Department CURRENCY EQUIVALENTS Currency Unit – yuan (CNY) At Appraisal At Project Completion At Operations Evaluation (July 1998) (August 2004) (December 2006) CNY1.00 = $0.1208 $0.1232 $0.1277 $1.00 = CNY8.28 CNY8.12 CNY7.83 ABBREVIATIONS AADT – annual average daily traffic ADB – Asian Development Bank CDB – China Development Bank DMF – design and monitoring framework EIA – environmental impact assessment EIRR – economic internal rate of return FIRR – financial internal rate of return GDP – gross domestic product ha – hectare HHEC – Heilongjiang Hashuang Expressway Corporation HPCD – Heilongjiang Provincial Communications Department ICB – international competitive bidding JPCD – Jilin Provincial Communications Department JPEC – Jilin Provincial Expressway Corporation MOC – Ministry of Communications NTHS – national trunk highway system O&M – operations and maintenance OEM – Operations Evaluation Mission PCD – provincial communication department PCR – project completion report PPTA – project preparatory technical assistance PRC – People’s Republic of China RRP – report and recommendation of the President TA – technical assistance VOC – vehicle operating cost NOTE In this report, “$” refers to US dollars. Keywords asian development bank, development effectiveness, expressways, people’s republic of china, performance evaluation, heilongjiang province, jilin province, transport Director Ramesh Adhikari, Operations Evaluation Division 2, OED Team leader Marco Gatti, Senior Evaluation Specialist, OED Team members Vivien Buhat-Ramos, Evaluation Officer, OED Anna Silverio, Operations Evaluation Assistant, OED Irene Garganta, Operations Evaluation Assistant, OED Operations Evaluation Department, PE-696 CONTENTS Page BASIC DATA v EXECUTIVE SUMMARY vii MAPS xi I. INTRODUCTION 1 A. -

Optimization Path of the Freight Channel of Heilongjiang Province
2017 3rd International Conference on Education and Social Development (ICESD 2017) ISBN: 978-1-60595-444-8 Optimization Path of the Freight Channel of Heilongjiang Province to Russia 1,a,* 2,b Jin-Ping ZHANG , Jia-Yi YUAN 1Harbin University of Commerce, Harbin, Heilongjiang, China 2International Department of Harbin No.9 High School, Harbin, Heilongjiang, China [email protected], [email protected] * Corresponding author Keywords: Heilongjiang Province, Russia, Freight Channel, Optimization Path. Abstract. Heilongjiang Province becomes the most important province for China's import and export trade to Russia due to its unique geographical advantages and strong complementary between industry and product structure. However, the existing problems in the trade freight channel layout and traffic capacity restrict the bilateral trade scale expansion and trade efficiency improvement. Therefore, the government should engage in rational distribution of cross-border trade channel, strengthen infrastructure construction in the border port cities and node cities, and improve the software support and the quality of service on the basis of full communication and coordination with the relevant Russian government, which may contribute to upgrade bilateral economic and trade cooperation. Introduction Heilongjiang Province is irreplaceable in China's trade with Russia because of its geographical advantages, a long history of economic and trade cooperation, and complementary in industry and product structures. Its total value of import and export trade to Russia account for more than 2/3 of the whole provinces and nearly 1/4 of that of China. After years of efforts, there exist both improvement in channel infrastructure, layout and docking and problems in channel size, functional positioning and layout, as well as important node construction which do not match with cross-border freight development. -

Organ Harvesting
Refugee Review Tribunal AUSTRALIA RRT RESEARCH RESPONSE Research Response Number: CHN31387 Country: China Date: 14 February 2007 Keywords: China – Heilongjiang – Harbin – Falun Gong – Organ harvesting This response was prepared by the Country Research Section of the Refugee Review Tribunal (RRT) after researching publicly accessible information currently available to the RRT within time constraints. This response is not, and does not purport to be, conclusive as to the merit of any particular claim to refugee status or asylum. Questions 1. Does No 1 Harbin hospital exist and have there been any reports or allegations of organ harvesting at that hospital? 2. Any reports or allegations of organ harvesting in A’chen District, Ha’erbin, Heilongjiang China 3.Any significant protests against organ harvesting in this part of China that they applicant may have attended or would know about? 4. Details of particular hospitals or areas where it has been alleged that organ harvesting is taking place 5. If the applicant has conducted ‘research’ what sort of things might he know about? 6. Any prominent people or reports related to this topic that the applicant may be aware of. 7. Anything else of relevance. RESPONSE 1. Does No 1 Harbin hospital exist and have there been any reports or allegations of organ harvesting at that hospital? Sources indicate that ‘No 1 Harbin Hospital’ does exist. References also mention a No 1 Harbin Hospital that is affiliated with Harbin Medical University. No reports regarding organ harvesting at No 1 Harbin Hospital where found in the sources consulted. Falun Gong sources have however provided reports alleging organ harvesting activities within No.1 Hospital Affiliated to Harbin Medical School. -

Investigation of Borrelia Spp. in Ticks (Acari: Ixodidae)
Asian Pacific Journal of Tropical Medicine (2012)459-464 459 Contents lists available at ScienceDirect Asian Pacific Journal of Tropical Medicine journal homepage:www.elsevier.com/locate/apjtm Document heading doi: Investigation of Borrelia spp. in ticks (Acari: Ixodidae) at the border crossings between China and Russia in Heilongjiang Province, China Shi Liu1, Chao Yuan2, Yun-Fu Cui1*, Bai-Xiang Li3, Li-Jie Wu3, Ying Liu4 1The 2nd Affiliated Hospital of Harbin Medical University, Harbin 150001, People s Republic of China 2 ' Daqing Oilfield General Hospital Group Rangbei Hospital, Daqing, 163114, People s Republic of China 3 ' Harbin Medical University School of Public Health, Harbin 150001, People s Republic of China 4 ' The 3rd Affiliated Hospital of Qiqihar Medical College, QiqiHar 161002, People s Republic of China ' ARTICLE INFO ABSTRACT Article history: Objective: Borrelia To investigate the precise species of tick vector andMethods: the spirochete pathogen Received 15 February 2012 at the Heilongjiang Province international border with Russia. In this study, ticks were Received in revised form 15 March 2012 collected from 12 Heilongjiang border crossings (including grasslands, shrublands, forests, and Accepted 15 April 2012 plantantions) to determine the rate and species type of spirochete-infected ticks and the most Available online 20 June 2012 Results: prevalent spirochete genotypes. The ticks represented three genera and four species Ixodes persulcatus Dermacentor silvarum Haemaphysalis concinna of the Ixodidae family [ , , and Haemaphysalis japonica Ixodes persulcatus Borrelia burgdorferi sensu ]. had the highest amount of Keywords: lato Borrelia Ixodes persulcatus infection of 25.6% and the most common species of isolated from Borrelia garinii Conclusions: Borrelia garinii Lyme disease was , strain PD91. -

Considering the Dynamic Dependence of Icing-Inducing Factors
Exploring the Highway Icing Risk: Considering the Dynamic Dependence of Icing-Inducing Factors Qiang Liu Harbin Institute of Technology Aiping Tang ( [email protected] ) Harbin Institute of Technology Zhongyue Wang Harbin Institute of Technology Buyue Zhao Beijing Jiaotong University Research Article Keywords: Dependence analysis, Exceeding probability, Hazard, Vulnerability, Transportation risk Posted Date: April 22nd, 2021 DOI: https://doi.org/10.21203/rs.3.rs-445530/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License 1 Exploring the highway icing risk: Considering the dynamic 2 dependence of icing-inducing factors 3 Qiang Liu a, Aiping Tang a, b *, Zhongyue Wang a,Buyue Zhao c 4 a School of Civil Engineering, Harbin Institute of Technology, Harbin 150090, China 5 b Key Lab of Smart Prevention and Mitigation of Civil Engineering Disasters of the Ministry of Industry and Information Technology, 6 Harbin Institute of Technology, Harbin150090, China 7 c School of Civil Engineering, Beijing Jiaotong University, Beijing 100044, China 8 Abstract:In terms of the dynamic dependence between icing-inducing factors, this study is to explore the risk 9 distribution of highways when icing events occur in the study area. A joint distribution considering the dynamic 10 correlation of inducing factors was first constructed employing the Copula theory, which then yielded the 11 possibility of icing events. Meanwhile, hazard zones and intensities of icing were proposed under different 12 exceeding probabilities. After finishing the vulnerability analysis of highways, the risk matrix was used to conduct 13 the icing risk for the highway, which was then applied to the construction of the risk zoning map. -

Annual Report 2019
HAITONG SECURITIES CO., LTD. 海通證券股份有限公司 Annual Report 2019 2019 年度報告 2019 年度報告 Annual Report CONTENTS Section I DEFINITIONS AND MATERIAL RISK WARNINGS 4 Section II COMPANY PROFILE AND KEY FINANCIAL INDICATORS 8 Section III SUMMARY OF THE COMPANY’S BUSINESS 25 Section IV REPORT OF THE BOARD OF DIRECTORS 33 Section V SIGNIFICANT EVENTS 85 Section VI CHANGES IN ORDINARY SHARES AND PARTICULARS ABOUT SHAREHOLDERS 123 Section VII PREFERENCE SHARES 134 Section VIII DIRECTORS, SUPERVISORS, SENIOR MANAGEMENT AND EMPLOYEES 135 Section IX CORPORATE GOVERNANCE 191 Section X CORPORATE BONDS 233 Section XI FINANCIAL REPORT 242 Section XII DOCUMENTS AVAILABLE FOR INSPECTION 243 Section XIII INFORMATION DISCLOSURES OF SECURITIES COMPANY 244 IMPORTANT NOTICE The Board, the Supervisory Committee, Directors, Supervisors and senior management of the Company warrant the truthfulness, accuracy and completeness of contents of this annual report (the “Report”) and that there is no false representation, misleading statement contained herein or material omission from this Report, for which they will assume joint and several liabilities. This Report was considered and approved at the seventh meeting of the seventh session of the Board. All the Directors of the Company attended the Board meeting. None of the Directors or Supervisors has made any objection to this Report. Deloitte Touche Tohmatsu (Deloitte Touche Tohmatsu and Deloitte Touche Tohmatsu Certified Public Accountants LLP (Special General Partnership)) have audited the annual financial reports of the Company prepared in accordance with PRC GAAP and IFRS respectively, and issued a standard and unqualified audit report of the Company. All financial data in this Report are denominated in RMB unless otherwise indicated. -

3 Òèõîîêåàíñêàß Ãåîëîãèß, 2015, Òîì 34, № 1, Ñ. 3–12 Zhang
ÈÕÎÎÊÅÀÍÑÊÀß ÃÅÎËÎÃÈß, 2015, òîì 34, 1, ñ. 3–12 УДК 551.73[551.781.33]:(510) LATE PALEOZOIC-EARLY MESOZOIC TECTONIC EVOLUTION IN THE EAST MARGIN OF THE JIAMUSI MASSIF, EASTERN NORTHEASTERN CHINA Zhang XingZhou, Guo Ye, Zhou JianBo, Zeng Zhen, Pu JianBin, Fu QiuLin College of Earth Sciences, Jilin University, Changchun 130061, Jilin, China, e-mail: [email protected] Поступила в редакцию 7 июля 2014 г. The Jiamusi massif is a major tectonic unit in the eastern part of NE China and composed chiefl y of the Early Paleozoic (about 500 Ma) metamorphosed crystalline basement containing Precambrian, even Archean crust and three suites of unmetamorphosed continental marginal sedimentary formations of the Devonian-Lower Carboniferous, the Late Carboniferous- Permian and the Late Triassic, which are similar to those in the Bureya and Khanka massifs in Russia. In the Devonian-Lower Carboniferous, the microcontinent consisting of the Jiamusi and Songnen massifs in China and the Bureya and Khanka massifs in Russia evolved independently, the eastern part of which was a passive continental margin, where a suite of the marine sedimentary-volcanic formation is overlain unconformably on the crystalline basement. The regional stratigraphic break in the middle Carboniferous in the whole northeastern China was related to the collision of the microcontinent with the Argun- Hinggan microcontinent in the west, indicating the formation of a new amalgamated continent (Heilongjiang plate). Therefore, the Late Carboniferous-Permian volcanic-sedimentary formation is the fi rst unitary cover on the Heilongjiang plate. The Late Carboniferous-Permian and the Late Triassic sedimentary formations in the eastern part of the Jiamusi-Bureya-Khanka microcontinent represent the evolutional features of the eastern continental margin of the Heilongjiang plate. -

Pore-Fractures of Coalbed Methane Reservoir Restricted by Coal Facies in Sangjiang-Muling Coal-Bearing Basins, Northeast China
energies Article Pore-Fractures of Coalbed Methane Reservoir Restricted by Coal Facies in Sangjiang-Muling Coal-Bearing Basins, Northeast China Yuejian Lu 1,2 , Dameng Liu 1,2,*, Yidong Cai 1,2, Qian Li 1,2 and Qifeng Jia 1,2 1 School of Energy Resources, China University of Geosciences, Beijing 100083, China; [email protected] (Y.L.); [email protected] (Y.C.); [email protected] (Q.L.); [email protected] (Q.J.) 2 Coal Reservoir Laboratory of National Engineering Research Center of Coalbed Methane Development & Utilization, Beijing 100083, China * Correspondence: [email protected]; Tel.: +86-10-82323971 Received: 17 January 2020; Accepted: 4 March 2020; Published: 5 March 2020 Abstract: The pore-fractures network plays a key role in coalbed methane (CBM) accumulation and production, while the impacts of coal facies on the pore-fractures network performance are still poorly understood. In this work, the research on the pore-fracture occurrence of 38 collected coals from Sangjiang-Muling coal-bearing basins with multiple techniques, including mercury intrusion porosimetry (MIP), micro-organic quantitative analysis, and optic microscopy, and its variation controlling of coal face were studied. The MIP curves of 38 selected coals, indicating pore structures, were subdivided into three typical types, including type I of predominant micropores, type II of predominant micropores and macropores with good connectivity, and type III of predominant micropores and macropores with poor connectivity. For coal facies, three various coal facies were distinguished, including lake shore coastal wet forest swamp, the upper delta plain wet forest swamp, tidal flat wet forest swamp using Q-cluster analysis and tissue preservation index–gelification index (TPI-GI), and wood index–groundwater influence index (WI-GWI). -

Palaeontology and Biostratigraphy of the Lower Cretaceous Qihulin
Dissertation Submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany for the degree of Doctor of Natural Sciences presented by Master of Science: Gang Li Born in: Heilongjiang, China Oral examination: 30 November 2001 Gedruckt mit Unterstützung des Deutschen Akademischen Austauschdienstes (Printed with the support of German Academic Exchange Service) Palaeontology and biostratigraphy of the Lower Cretaceous Qihulin Formation in eastern Heilongjiang, northeastern China Referees: Prof. Dr. Peter Bengtson Prof. Pei-ji Chen This manuscript is produced only for examination as a doctoral dissertation and is not intended as a permanent scientific record. It is therefore not a publication in the sense of the International Code of Zoological Nomenclature. Abstract The purpose of the study was to provide conclusive evidence for a chronostratigraphical assignment of the Qihulin Formation of the Longzhaogou Group exposed in Mishan and Hulin counties of eastern Heilongjiang, northeastern China. To develop an integrated view of the formation, all collected fossil groups, i.e. the macrofossils (ammonites and bivalves) and microfossils (agglutinated foraminifers and radiolarians) have been studied. The low-diversity ammonite fauna consists of Pseudohaploceras Hyatt, 1900, and Eogaudryceras Spath, 1927, which indicate a Barremian–Aptian age. The bivalve fauna consists of eight genera and 16 species. The occurrence of Thracia rotundata (J. de C: Sowerby) suggests an Aptian age. The agglutinated foraminifers comprise ten genera and 16 species, including common Lower Cretaceous species such as Ammodiscus rotalarius Loeblich & Tappan, 1949, Cribrostomoides? nonioninoides (Reuss, 1836), Haplophragmoides concavus (Chapman, 1892), Trochommina depressa Lozo, 1944. The radiolarians comprise ten genera and 17 species, where Novixitus sp., Xitus cf. -
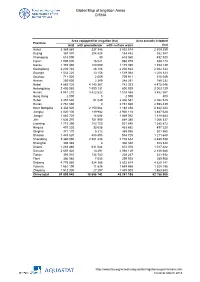
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52 -

Heilongjiang Road Development II Project (Yichun-Nenjiang)
Technical Assistance Consultant’s Report Project Number: TA 7117 – PRC October 2009 People’s Republic of China: Heilongjiang Road Development II Project (Yichun-Nenjiang) FINAL REPORT (Volume II of IV) Submitted by: H & J, INC. Beijing International Center, Tower 3, Suite 1707, Beijing 100026 US Headquarters: 6265 Sheridan Drive, Suite 212, Buffalo, NY 14221 In association with WINLOT No 11 An Wai Avenue, Huafu Garden B-503, Beijing 100011 This consultant’s report does not necessarily reflect the views of ADB or the Government concerned, ADB and the Government cannot be held liable for its contents. All views expressed herein may not be incorporated into the proposed project’s design. Asian Development Bank Heilongjiang Road Development II (TA 7117 – PRC) Final Report Supplementary Appendix A Financial Analysis and Projections_SF1 S App A - 1 Heilongjiang Road Development II (TA 7117 – PRC) Final Report SUPPLEMENTARY APPENDIX SF1 FINANCIAL ANALYSIS AND PROJECTIONS A. Introduction 1. Financial projections and analysis have been prepared in accordance with the 2005 edition of the Guidelines for the Financial Governance and Management of Investment Projects Financed by the Asian Development Bank. The Guidelines cover both revenue earning and non revenue earning projects. Project roads include expressways, Class I and Class II roads. All will be built by the Heilongjiang Provincial Communications Department (HPCD). When the project started it was assumed that all project roads would be revenue earning. It was then discovered that national guidance was that Class 2 roads should be toll free. The ADB agreed that the DFR should concentrate on the revenue earning Expressway and Class I roads, 2.