A Component of Retinal Light Adaptation Mediated by the Thyroid Hormone Cascade
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
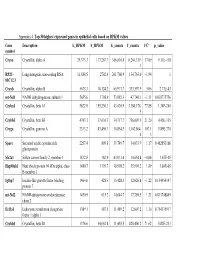
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

CRYGB Rabbit Pab
Leader in Biomolecular Solutions for Life Science CRYGB Rabbit pAb Catalog No.: A14569 Basic Information Background Catalog No. Crystallins are separated into two classes: taxon-specific, or enzyme, and ubiquitous. A14569 The latter class constitutes the major proteins of vertebrate eye lens and maintains the transparency and refractive index of the lens. Since lens central fiber cells lose their Observed MW nuclei during development, these crystallins are made and then retained throughout 21kDa life, making them extremely stable proteins. Mammalian lens crystallins are divided into alpha, beta, and gamma families; beta and gamma crystallins are also considered as a Calculated MW superfamily. Alpha and beta families are further divided into acidic and basic groups. 20kDa Seven protein regions exist in crystallins: four homologous motifs, a connecting peptide, and N- and C-terminal extensions. Gamma-crystallins are a homogeneous group of Category highly symmetrical, monomeric proteins typically lacking connecting peptides and terminal extensions. They are differentially regulated after early development. Four Primary antibody gamma-crystallin genes (gamma-A through gamma-D) and three pseudogenes (gamma-E, gamma-F, gamma-G) are tandemly organized in a genomic segment as a Applications gene cluster. Whether due to aging or mutations in specific genes, gamma-crystallins WB have been involved in cataract formation. Cross-Reactivity Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 1419 P07316 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 80-135 of human CRYGB (NP_005201.2). Synonyms CRYGB;CRYG2;CTRCT39 Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. -

A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi Washington University School of Medicine in St. Louis Rafal Farjo University of Michigan - Ann Arbor Shigeo Yoshida University of Michigan - Ann Arbor Timothy S. Kern Case Western Reserve University Anand Swaroop University of Michigan - Ann Arbor See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Xi, Jinghua; Farjo, Rafal; Yoshida, Shigeo; Kern, Timothy S.; Swaroop, Anand; and Andley, Usha P., ,"A comprehensive analysis of the expression of crystallins in mouse retina." Molecular Vision.9,. 410-419. (2003). https://digitalcommons.wustl.edu/open_access_pubs/1801 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jinghua Xi, Rafal Farjo, Shigeo Yoshida, Timothy S. Kern, Anand Swaroop, and Usha P. Andley This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/1801 Molecular Vision 2003; 9:410-9 <http://www.molvis.org/molvis/v9/a53> © 2003 Molecular Vision Received 28 May 2003 | Accepted 19 August 2003 | Published 28 August 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi,1 Rafal Farjo,3 Shigeo Yoshida,3 Timothy S. Kern,5 Anand Swaroop,3,4 Usha P. Andley1,2 Departments of 1Ophthalmology and Visual Sciences and 2Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. -

Related Macular Degeneration and Cutis Laxa
UvA-DARE (Digital Academic Repository) Genetic studies of age-related macular degeneration Baas, D.C. Publication date 2012 Document Version Final published version Link to publication Citation for published version (APA): Baas, D. C. (2012). Genetic studies of age-related macular degeneration. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:05 Oct 2021 G������ S������ �� A��-������� M������ D����������� D����������� M������ G������ S������ �� A��-������� | 2012 D�������� C. B��� G������ S������ �� A��-������� M������ D����������� D�������� C. B��� cover.indd 1 31-10-12 08:36 Genetic Studies of Age-related Macular Degeneration Dominique C. Baas Chapter 0.indd 1 23-10-12 19:24 The research described in this thesis was conducted at the Netherlands Institute for Neuroscience (NIN), an institute of the Royal Netherlands Academy of Arts and Sciences, Department of Clinical and Molecular Ophthalmogenetics, Amsterdam, The Netherlands. -

Congenital Cataracts Due to a Novel 2‑Bp Deletion in CRYBA1/A3
1614 MOLECULAR MEDICINE REPORTS 10: 1614-1618, 2014 Congenital cataracts due to a novel 2‑bp deletion in CRYBA1/A3 JING ZHANG1, YANHUA ZHANG1, FANG FANG1, WEIHONG MU1, NING ZHANG2, TONGSHUN XU3 and QINYING CAO1 1Prenatal Diagnosis Center, Shijiazhuang Obstetrics and Gynecology Hospital; 2Department of Cardiology, The Second Hospital of Hebei Medical University; 3Department of Surgery, Shijiazhuang Obstetrics and Gynecology Hospital, Shijiazhuang, Hebei, P.R. China Received September 22, 2013; Accepted April 11, 2014 DOI: 10.3892/mmr.2014.2324 Abstract. Congenital cataracts, which are a clinically and located in the eye lens. The major human crystallins comprise genetically heterogeneous group of eye disorders, lead to 90% of protein in the mature lens and contain two different visual impairment and are a significant cause of blindness superfamilies: the small heat‑shock proteins (α-crystallins) in childhood. A major proportion of the causative mutations and the βγ-crystallins. for congenital cataracts are found in crystallin genes. In the In this study a functional candidate approach was used present study, a novel deletion mutation (c.590-591delAG) in to investigate the known crystallin genes, including CRYAA, exon 6 of CRYBA1/A3 was identified in a large family with CRYAB, CRYBA1/A3, CRYBB1, CRYBB2, CRYGC, CRYGD autosomal dominant congenital cataracts. An increase in and CRYGS, in which a major proportion of the mutations local hydrophobicity was predicted around the mutation site; identified in a large family with congenital cataracts were however, further studies are required to determine the exact found. effect of the mutation on βA1/A3-crystallin structure and function. To the best of our knowledge, this is the first report Subjects and methods of an association between a frameshift mutation in exon 6 of CRYBA1/A3 and congenital cataracts. -

Congenital Eye Disorders Gene Panel
Congenital eye disorders gene panel Contact details Introduction Regional Genetics Service Ocular conditions are highly heterogeneous and show considerable phenotypic overlap. 1 in Levels 4-6, Barclay House 2,500 children in the UK are diagnosed as blind or severely visually impaired by the time they 37 Queen Square reach one year old. As many as half of these cases are likely to be inherited and remain undiagnosed due to the vast number of genes involved in these conditions. Many congenital London, WC1N 3BH eye disorders causing visual impairment or blindness at birth or progressive visual impairment T +44 (0) 20 7762 6888 also include syndromic conditions involving additional metabolic, developmental, physical or F +44 (0) 20 7813 8578 sensory abnormalities. Gene panels offer the enhanced probability of diagnosis as a very large number of genes can be interrogated. Samples required Ocular birth defects include all inheritance modalities. Autosomal dominant and recessive 5ml venous blood in plastic EDTA diseases as well as X-linked dominant and recessive diseases are seen. These conditions can bottles (>1ml from neonates) also be caused by de novo variants. Prenatal testing must be arranged Referrals in advance, through a Clinical Genetics department if possible. Patients presenting with a phenotype appropriate for the requested sub-panel Amniotic fluid or CV samples Referrals will be accepted from clinical geneticists and consultants in ophthalmology. should be sent to Cytogenetics for Prenatal testing dissecting and culturing, with instructions to forward the sample Prenatal diagnosis may be offered as appropriate where pathogenic variants have been to the Regional Molecular Genetics identified in accordance with expected inheritance pattern and where appropriate parental laboratory for analysis testing and counselling has been conducted. -

Supplementary Materials
Supplementary materials Gene Nucleotide Amino acid change Clinical phenotype Ref symbol change CRYAA c.35G>T p. R12L lens protein gene [1] CRYAB c.32G>A p. R11H lens protein gene [2] CRYBA1 c.279-281delG p.ΔG91 lens protein gene [3] AG CRYBA4 c.206T>C p. L69P lens protein gene [4] CRYBB1 c.658G>T p. G220X lens protein gene [5] CRYBB2 c.563G>A p. R188H lens protein gene [6] CRYBB3 c.314G>A p. R105Q lens protein gene [7] CRYGA c.196T>C p. Y66H lens protein gene [8] CRYGB c.449G>T p. G150V lens protein gene [8] CRYGC c.385G>T p. G129C lens protein gene [9] CRYGD c.70C>A p. P24T lens protein gene [10] CRYGS c.53G>T :p.G18V lens protein gene [11] GJA3 c.188A>G p.N63S membrane protein [12] gene GJA8 c.262C>T p.P88S membrane protein [13] gene BFSP1 c736-1384_c.9 T246fsX7 cytoskeleton protein [14] 57-66del gene BFSP2 c.1091G>A p.G364D cytoskeleton protein [15] gene PAX6 c.307C>T p.R103X developmental [16] regulatory protein gen PITX3 c.38G>A p.S13N developmental [17] regulatory protein gen HSF4 c.524G>C p.R175P developmental [18] regulatory protein gen MAF c.863G>C p.R288P developmental [19] regulatory protein gen CHMP4B c.481G>A p. E161K chromatin modified [20] protein gene EPHA2 c.2842G>T p. G948W tyrosine kinase [21] receptor gene COL4A1 c.2345G>C p. G782A syndrome-related [22] genes FTL c.160G>A p.E54K developmental [23] regulatory protein gen GALK1 c.416T>C p. -

A Novel Γd-Crystallin Mutation Causes Mild Changes in Protein Properties but Leads to Congenital Coralliform Cataract
Molecular Vision 2009; 15:1521-1529 <http://www.molvis.org/molvis/v15/a162> © 2009 Molecular Vision Received 29 April 2009 | Accepted 3 August 2009 | Published 6 August 2009 A novel γD-crystallin mutation causes mild changes in protein properties but leads to congenital coralliform cataract Li-Yun Zhang,1 Bo Gong,1 Jian-Ping Tong,2 Dorothy Shu-Ping Fan,1 Sylvia Wai-Yee Chiang,1 Dinghua Lou,2 Dennis Shun-Chiu Lam,1 Gary Hin-Fai Yam,1 Chi-Pui Pang1 1Department of Ophthalmology and Visual Sciences, The Chinese University of Hong Kong, Hong Kong, China; 2Department of Ophthalmology, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China Purpose: To identify the genetic lesions for congenital coralliform cataract. Methods: Two Chinese families with autosomal dominant coralliform cataract, 12 affected and 14 unaffected individuals, were recruited. Fifteen known genes associated with autosomal dominant congenital cataract were screened by two-point linkage analysis with gene based single nucleotide polymorphisms and microsatellite markers. Sequence variations were identified. Recombinant FLAG-tagged wild type or mutant γD-crystallin was expressed in human lens epithelial cells and COS-7 cells. Protein solubility and intracellular distribution were analyzed by western blotting and immunofluorescence, respectively. Results: A novel heterozygous change, c.43C>A (R15S) of γD-crystallin (CRYGD) co-segregated with coralliform cataract in one family and a known substitution, c.70C>A (P24T), in the other family. Unaffected family members and 103 unrelated control subjects did not carry these mutations. Similar to the wild type protein, R15S γD-crystallin was detergent soluble and was located in the cytoplasm. -

A Recurrent Mutation in CRYGD Is Associated with Autosomal Dominant Congenital Coralliform Cataract in Two Unrelated Chinese Families
Molecular Vision 2011; 17:1085-1089 <http://www.molvis.org/molvis/v17/a123> © 2011 Molecular Vision Received 29 March 2011 | Accepted 19 April 2011 | Published 28 April 2011 A recurrent mutation in CRYGD is associated with autosomal dominant congenital coralliform cataract in two unrelated Chinese families Guoxing Yang,1 Chunlei Xiong,2 Shanlan Li,2 Yuanyuan Wang,2 Jialiang Zhao1 1Department of Opthalmology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China; 2Department of Opthalmology, Hai Cheng Rehabilitation Hospital, Liaoning, China Purpose: Congenital cataract is a clinically and genetically heterogeneous lens disorder. The purpose of this study was to identify the mutation responsible for autosomal dominant congenital coralliform cataracts in two Chinese families and to investigate the relationship between virulence genes and lens morphology. Methods: Patients received a physical examination, and blood samples were collected for DNA extraction. Mutation analysis was performed by direct sequencing of the candidate genes: gammaC-crystallin (CRYGC), gammaD-crystallin (CRYGD), gammaS-crystallin (CRYGS), gap-junction protein, alpha 8 (GJA8), gap-junction protein, alpha 3 (GJA3), and alphaA-crystallin (CRYAA). Results: The affected individuals in two families had congenital coralliform cataracts. Mutational analysis of the CRYGD identified a C→A transversion at nucleotide position c.70 in exon 2, which resulted in a threonine substitution for proline at amino-acid residue 24 (P24T). This mutation was identified in all affected individuals but was not found in healthy relatives or 100 control chromosomes from the same ethnic background. Conclusions: Our results indicated that the P24T mutation of CRYGD was responsible for two Chinese pedigrees with congenital coralliform cataracts. -

V13a25-Kamphuis Pgmkr
Molecular Vision 2007; 13:220-8 <http://www.molvis.org/molvis/v13/a25/> ©2007 Molecular Vision Received 30 October 2006 | Accepted 27 January 2007 | Published 8 February 2007 Transfer of lens-specific transcripts to retinal RNA samples may underlie observed changes in crystallin-gene transcript levels after ischemia Willem Kamphuis,1,2 Frederike Dijk,1 Willem Kraan,1 Arthur A.B. Bergen1 1Department of Molecular Ophthalmogenetics, 2Department of Cellular Quality Control, Netherlands Institute for Neuroscience- KNAW, Meibergdreef 47, 1105 BA Amsterdam, The Netherlands Purpose: Retinal ischemia appears to lead to alterations in retinal transcript levels of a group of genes known to be abundantly expressed in the lens. Our purpose is to study whether these alterations are truly the result of retinal ischemia or whether they could be caused by contamination of the retinal tissue with trace amounts of lens tissue. Methods: Changes occurring in the retinal gene expression profile after induction of retinal ischemia were assessed by oligonucleotide microarrays and by real-time quantitative PCR. Results: Microarray analysis of the retinal gene expression profile after 5 or 60 min ischemia showed altered transcript levels for a group of genes with functions related to “structural constituent of eye lens” (23 genes, predominantly crystallins). Subsequent qPCR assays for this set of genes showed extremely high variations in transcript levels between individual animals of both control and ischemia-treated groups. However, the relative transcript levels, or expression profile, of these genes was constant in all samples. The transcript levels of these genes were on average 2624-times higher in tissue samples isolated from the superficial layers of the total lens. -

Crystallin, Γ-Crystallin and GJA8 Gene in Congenital Cataract
Molecular Vision 2011; 17:693-707 <http://www.molvis.org/molvis/v17/a79> © 2011 Molecular Vision Received 10 October 2010 | Accepted 7 March 2011 | Published 11 March 2011 Mutation screening and genotype phenotype correlation of α- crystallin, γ-crystallin and GJA8 gene in congenital cataract Manoj Kumar,1 Tushar Agarwal,2 Sudarshan Khokhar,2 Manoj Kumar,3 Punit Kaur,3 Tara Sankar Roy,4 Rima Dada1 1Laboratory for Molecular Reproduction and Genetics, Department of Anatomy, All India Institute of Medical Sciences, New Delhi, India; 2Dr. Rajendra Prasad Centre for Ophthalmic Sciences, All India Institute of Medical Sciences, New Delhi, India; 3Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India; 4Department of Anatomy, All India Institute of Medical Sciences, New Delhi, India Purpose: To screen α-crystallin (CRYAB), γ-crystallin (CRYGC and CRYGD), and Connexin 50 (Cx-50 or GJA8) genes in congenital cataract patients and controls. Methods: Thirty clinically diagnosed congenital cataract cases below 3 years of age from northern India, presenting at Dr. R. P. Centre for Ophthalmic Sciences (AIIMS, New Delhi, India) were enrolled in this study. Genomic DNA was extracted from peripheral blood, all coding and exon/intron regions were amplified using PCR and direct sequencing was performed to detect any nucleotide variation. ProtScale and Discovery Studio programs were used for insilico and structural analysis of non-synonymous mutations. Results: DNA sequencing analysis of CRYAB, CRYGC, CRYGD, and GJA8 showed a total of six variations of which two were novel (CRYGC:p.R48H and GJA8:p.L281C) and four have been previously reported (CRYAB: rs11603779T>G, GJA8: p.L268L, CRYGD: p.R95R, and c.T564C). -

Zebrafish Model in Ophthalmology to Study Disease Mechanism And
pharmaceuticals Review Zebrafish Model in Ophthalmology to Study Disease Mechanism and Drug Discovery Yiwen Hong and Yan Luo * State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-Sen University, Guangzhou 510060, China; [email protected] * Correspondence: [email protected]; Tel.: +86-020-87335931 Abstract: Visual impairment and blindness are common and seriously affect people’s work and quality of life in the world. Therefore, the effective therapies for eye diseases are of high priority. Zebrafish (Danio rerio) is an alternative vertebrate model as a useful tool for the mechanism elucidation and drug discovery of various eye disorders, such as cataracts, glaucoma, diabetic retinopathy, age-related macular degeneration, photoreceptor degeneration, etc. The genetic and embryonic accessibility of zebrafish in combination with a behavioral assessment of visual function has made it a very popular model in ophthalmology. Zebrafish has also been widely used in ocular drug discovery, such as the screening of new anti-angiogenic compounds or neuroprotective drugs, and the oculotoxicity test. In this review, we summarized the applications of zebrafish as the models of eye disorders to study disease mechanism and investigate novel drug treatments. Keywords: zebrafish; eye; disease model; mechanism; drug candidate Citation: Hong, Y.; Luo, Y. Zebrafish Model in Ophthalmology to Study 1. Introduction Disease Mechanism and Drug Until 2020, an estimated 295 million people suffer from the moderate and severe visual Discovery. Pharmaceuticals 2021, 14, impairment worldwide, and among them, about 43.3 million people are even blind [1]. 716. https://doi.org/10.3390/ The leading global causes of blindness are cataract, followed by glaucoma, age-related ph14080716 macular degeneration (AMD), and diabetic retinopathy (DR) [2].