Introgression Between Pinus Taeda L. and Pinus Echinata Mill
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hwd”Rni -I I Southern Rest SO-296 Experimen?” Station July, 1963
c=.. United States I &=*$ Departmenlt of ‘$!j’!&’ Argriculture Forest Service hWD”rnI -I I Southern rest SO-296 Experimen?” Station July, 1963 Lightning Strike Simulation for Studying Southern Pine Bark and Engraver Beetle Attacks Mitchel C. Miller SUMMARY Endemic populations of the southern pine beetle is probably conservative; actual values could be (Dendroctonus frontalis Zimm.) and Ips spp. at- considerably higher. tacked loblolly pilnes (Pinus taeda L.) on which light- Lightning struck trees may attract SPB with a re- ning strikes were simulated with detonating cord lease of volatile oleoresin fractions from the shower in the field. Southern pine beetles were reared in of finely divided bark and needle particles (Taylor successive generations in these trees from fall 1981 1973, Lorio and Yandle 1978); struck trees offer a through spring 1982; only Ips spp. attacked treated favorable attack and brood development site for trees through December 1982. Lightning strike SPB and Ips spp. as a result of reduced oleoresin simulation provides an alternative means of studying exudation pressure, reduced oleoresin flow, re- bark beetle attack and opens the way for com- duced relative water content and increased reduc- parisons with nat;ural lightning strikes. ing-sugar content of inner bark (Hodges and Pickard Keywords: Lightning strike simulation, southern 1971). Anderson and Anderson (1968) associated pine beetle, Ips, Dendroctonus frontalis. successful attack by three Ips species on a light- ning struck loblolly pine with reduced oleoresin -

Native Woody Plants of Montgomery County, Maryland
Native Woody Plants of Montgomery County, Maryland ~ John Mills Parrish, 2002 Plant List State Where Latin Name Common Name Rank/Status Occurrence Found GYMNOSPERMAE - GYMNOSPERMS Cupressaceae - Cypress Family Juniperus Red Cedar C virginiana Pinaceae - Pine Family Pinus strobus White Pine VR Patuxent St. Park; Northwest Br. Park Pinus rigida Pitch Pine UC Scattered throughout county Pinus echinata Yellow/Shortleaf Pine UC Scattered throughout county Pinus pungens Table-mountain Pine VR NW Branch Pk; Blockhouse Pt. Park Pinus Virginia Pine C virginiana Tsuga Hemlock VR Patuxent St. canadensis Pk; Seneca Ck. St. Park ANGIOSPERMAE - MONOCOTS Smilacaceae - Catbrier Family Smilax glauca Glaucous Greenbrier C Smilax hispida Bristly Greenbrier UC/R Potomac (syn. S. River & Rock tamnoides) Ck. floodplain Smilax Common Greenbrier C rotundifolia ANGIOSPERMAE - DICOTS Salicaceae - Willow Family Salix nigra Black Willow C Salix Carolina Willow S3 R Potomac caroliniana River floodplain Salix interior Sandbar Willow S1/E VR/X? Plummer's & (syn. S. exigua) High Is. (1902) (S.I.) Salix humilis Prairie Willow R Travilah Serpentine Barrens Salix sericea Silky Willow UC Little Bennett Pk.; NW Br. Pk. (Layhill) Populus Big-tooth Aspen UC Scattered grandidentata across county - (uplands) Populus Cottonwood FC deltoides Myricaceae - Bayberry Family Myrica cerifera Southern Bayberry VR Little Paint Branch n. of Fairland Park Comptonia Sweet Fern VR/X? Lewisdale, peregrina (pers. com. C. Bergmann) Juglandaceae - Walnut Family Juglans cinerea Butternut S2S3 R -

Pinus Echinata Shortleaf Pine
PinusPinus echinataechinata shortleafshortleaf pinepine by Dr. Kim D. Coder, Professor of Tree Biology & Health Care Warnell School of Forestry & Natural Resources, University of Georgia One of the most widespread pines of the Eastern United Sates is Pinus echinata, shortleaf pine. Shortleaf pine was identified and named in 1768. The scientific name means a “prickly pine cone tree.” Other common names for shortleaf pine include shortstraw pine, yellow pine, Southern yellow pine, shortleaf yellow pine, Arkansas soft pine, Arkansas pine, and old field pine. Among all the Southern yellow pines it has the greatest range and is most tolerant of a variety of sites. Shortleaf pine grows Southeast of a line between New York and Texas. It is widespread in Georgia except for coastal coun- ties. Note the Georgia range map figure. Pinus echinata is found growing in many mixtures with other pines and hardwoods. It tends to grow on medium to dry, well-drained, infertile sites, as compared with loblolly pine (Pinus taeda). It grows quickly in deep, well-drained areas of floodplains, but cannot tolerate high pH and high calcium concentrations. Compared with other Southern yellow pines, shortleaf is less demanding of soil oxygen content and essential element availability. It grows in Hardiness Zone 6a - 8b and Heat Zone 6-9. The lowest number of Hardiness Zone tends to delineate the Northern range limit and the largest Heat Zone number tends to define the South- ern edge of the range. This native Georgia pine grows in Coder Tree Grow Zone (CTGZ) A-D (a mul- tiple climatic attribute based map), and in the temperature and precipitation cluster based Coder Tree Planting Zone 1-6. -

Overcoming Dormancy in Loblolly Pine (Pinus Taeda L.)
Reprinted from: Edwards, D.G.W., compiler and editor. 1993. "Dormancy and barriers to germination." Proc. Internal. Sympos. IUFRO Project Group P2.04-00 (Seed Problems). Victoria, British Columbia Canada ' April 23-26, 1991. Forestry Canada, Pacific Forestry Centre, Victoria, B.C. Overcoming dormancy in loblolly pine (Pinus taeda L.) F.T. BONNER AND C.A. HARRINGTON u.s. Department of Agriculture, Forest Service Southern Forest Experiment Station, Starkville, MS 39759, U.S.A. and Pacific Northwest Research Station, Olympia, WA 98502, U.S.A. Abstract Dormancy patterns in loblolly pine (Pinus taeda L.) were examined by comparing various parameters of germination rate. Both linear and polynomial models showed that mean germination time (MGT) was the most sensitive to chilling period. MGT was then used in a test of a new chilling technique that employed 24-h warm interruptions of chilling. This procedure produced faster rates of germination (lower MGT) in one group of 9 lots, but a second test with 15 lots yielded negative responses to any type of stratification. Resume Chez Ie pin a encens (Pinus taeda L.), les caracteristiques de la dormance ont ete examinees par comparaison de differents parametres du taux de germination. Des modeles lineaires comme polynomiaux ont montre que Ie temps moyen de germination etait Ie plus sensible a la periode d'entreposage en chambre froide. Cette duree moyenne a ensuite ete mise a profit dans l'essai d'une nouvelle technique d'entreposage en chambre froide qui comporte des interruptions de 24 h de rechauffement. Cette technique a accelere les taux de germination (temps moyen de germination raccourci) dans un groupe de 9 lots, mais lors d'un second essai sur 15 lots, a conduit a des resultats negatifs, peu importe Ie type de stratification. -

Tfsbbstudy.Pdf
Introduction Results Pine bark beetles, including the mountain pine beetle (MPB), Table 1. Effects of abamectin (Aba) injection treatments on the success of Ips engraver beetle adult Trial 1: Evaluation of bolts collected from treated and untreated Dendroctonus ponderosae Hopkins, and southern pine beetle (SPB), D. attack, brood development, and success of cerambycid larvae in loblolly pine logs in Texas: 2008 - loblolly pine trees in 2008 - 2011 indicated that a similar number frontalis Zimmermann, are some of the more important forest pests in 2011. of Ips engraver beetles attacked the bolts regardless of treatment the United States (Billings et al. 2004) with local and regional outbreaks (Table 1). However, the success of the Ips in constructing galleries causing severe economic losses on a nearly annual basis. Other species (> 2.5 cm) or producing brood was significantly less for both of pine bark beetles, including the secondary pests Ips avulsus (Eichoff), injection treatments, regardless of season, compared to the I. grandicollis (Eichoff), and I. calligraphus (Germar), also are known to checks (Fig. 3). Similarly, the number of cerambycid egg niches cause significant tree mortality particularly during severe drought periods was similar for all treatments, but there were significantly less in the southeastern U.S (Wilkinson & Foltz 1982). The current abundance phloem consumed by larvae in the injected logs. of susceptible trees and forests underlines the need to develop new methods to protect individual trees from bark beetle attacks. Trial 2: Nearly all baited trees were heavily attacked by MPB within 3 weeks. A final assessment for the 2010 season was Protection of individual trees with conducted in September 2011 and showed heavy mortality (60%, insecticides has historically 18 of 30) of untreated lodgepole pine trees (Fig. -
![SHORTLEAF PINE [Pinaceae] Pinus Echinata Miller](https://docslib.b-cdn.net/cover/6490/shortleaf-pine-pinaceae-pinus-echinata-miller-586490.webp)
SHORTLEAF PINE [Pinaceae] Pinus Echinata Miller
Vascular Plants of Williamson County Pinus echinata − SHORTLEAF PINE [Pinaceae] Pinus echinata Miller (possible selection, mixed-sized plants in woodland of escaped plants originating from cultivated specimens), SHORTLEAF PINE. Tree, evergreen, with 1 trunk to 30 cm diameter (non-cultivated), in range 4−10 m tall (reproductive); monoecious; shoots with long shoot-short shoot organization, long shoot growth beginning in late March, with closely spaced, nonphotosynthetic scale leaves along new axis (new spring growth after pollination begins), springtime long shoot initially to 200 × 8 mm before foliage leaf elongating, at each node having a scalelike primary leaf but immediately producing a short shoot in the axil of scale leaf (sylleptic development) = a “fascicle” of 2−3 photosynthetic leaves (needles) held together by several tightly wrapped papery (scarious) leaves at the base, glabrous, having resin ducts within plant, aromatic, especially when crushed or damaged. Stems: internodes relatively short hidden by persistent bases of scale leaves; young twigs flexible, lacking leaves when > 4 mm diameter, with helically arranged remnants of scale leaves; older stems with woody seed cones mostly 7−8 mm diameter shedding tannish gray leaf bases and forming light brown periderm; bark large- scaly, not peeling and tightly attached, gray, not obviously resinous. Leaves: of 3 types (scale, foliage, wrapper); scale leaves on new long shoots helically alternate, simple, sessile with decurrent bases; foliage leaves (needles) terminal and mostly -

Proceedings 33Rd Southern Forest Tree Improvement Conference Using Tree Improvement to Grow New Products for a Changing World Ho
Proceedings 33rd Southern Forest Tree Improvement Conference Using Tree Improvement to Grow New Products for a Changing World Hosted by: Arkansas Forest Resources Center; University of Arkansas Division of Agriculture School of Forest and Natural Resources; University of Arkansas at Monticello Weyerhaeuser Inc. Hot Springs, AR June 8-11, 2015 Proceedings of the 33rd Southern Forest Tree Improvement Conference Edited by Joshua P. Adams Louisiana Tech University Ruston, LA 71270 The papers and abstracts in these proceedings were submitted by the authors as electronic files. Modifications were made in format to provide for consistency and to allow best possible placement of figures and tables. The authors are responsible for the technical content of their respective papers. Citation for the 33rd SFTIC Proceedings: Adams, J.P., editor. 2016. Proceedings of the 33rd Southern Forest Tree Improvement Conference; 8-11 June 2015; Hot Springs, Arkansas. http://www.sftic.org 113 p. Citation of papers in the 33rd SFTIC Proceedings: Authors’ Names. 2016. Name of paper. In: Proceedings of the 33rd Southern Forest Tree Improvement Conference; 8-11 June 2015 Hot Springs, Arkansas. http://www.sftic.org , pp. x-y. Links to electronic copies may be obtained at: http://www.sftic.org ii Foreword The 33rd Southern Forest Tree Improvement Conference (SFTIC), marking 64 years of biennial technical conferences, was held June 8-11, 2015 in Hot Springs, Arkansas. This marks the first time the conference has been in the state of Arkansas. Direction of the conference was coordinated by the Southern Forest Tree Improvement Committee and with co-hosts Arkansas Forest Resources Center; University of Arkansas Division of Agriculture School of Forest and Natural Resources; University of Arkansas at Monticello and Weyerhaeuser Inc. -

Species Comparison of the Physical Properties of Loblolly and Slash Pine Wood and Bark Thomas L
1495 ARTICLE Species comparison of the physical properties of loblolly and slash pine wood and bark Thomas L. Eberhardt, Joseph Dahlen, and Laurence Schimleck Abstract: Composition of the southern pine forest is now predominated by two species, loblolly pine (Pinus taeda L.) and slash pine (Pinus elliottii Engelm.), owing to fire suppression activities, natural regeneration on abandoned agricultural lands, and extensive planting. Comparison of the wood and bark physical properties of these pines is of interest in terms of the yields of usable biomass and, for the bark, its ecological functionality on a living tree. Trees from a species comparison study were used to generate wood and bark property data, on a whole-tree basis, and for stem disks collected at breast height. Models were constructed to explain the effect of relative height on wood and bark properties. When comparing the whole-tree data, slash pine wood (0.523 versus 0.498) and bark (0.368 versus 0.311) specific gravity values were higher, both offset by lower moisture contents; slash pine also produced a higher percentage of bark on a dry-mass basis (17% versus 12.5%). Unlike wood properties, bark properties showed significant between-species differences when determined at breast height alone, the exception being moisture content. In terms of yield, harvests of a green tonne of loblolly pine and slash pine would give approximately the same dry mass of wood, but slash pine provides more bark. Key words: bark thickness, moisture content, specific gravity, wood quality, yield. Résumé : La composition de la forêt de pins du sud est maintenant dominée par deux espèces, le pin a` encens (Pinus taeda L.) et le pin d’Elliott (Pinus elliottii Engelm.) a` cause des activités de suppression des feux, de la régénération naturelle sur les terres agricoles abandonnées et de la plantation intensive. -

Pinus Echinata)-Hardwood Mixtures in Low Quality Mixed Upland Hardwood Stands Using Cluster Planting and Natural Regeneration
Article Restoration of Shortleaf Pine (Pinus echinata)-Hardwood Mixtures in Low Quality Mixed Upland Hardwood Stands Using Cluster Planting and Natural Regeneration David Clabo 1,* and Wayne Clatterbuck 2 1 Warnell School of Forestry & Natural Resources, University of Georgia, 4601 Research Way, Tifton, GA 31793, USA 2 Department of Forestry, Wildlife & Fisheries, University of Tennessee, 274 Ellington Plant Sciences Building, Knoxville, TN 37996, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-229-386-3672 Received: 3 April 2020; Accepted: 15 April 2020; Published: 17 April 2020 Abstract: Cluster planting of shortleaf pine, along with various site preparation and release treatments, were tested to restore mixed shortleaf pine (Pinus echinata Mill.)–hardwood stands in areas where the shortleaf pine has diminished in recent years. Shortleaf pine–hardwood mixtures were once a common forest type throughout the Cumberland Mountains and Plateau physiographic regions of the southeastern United States. Knowledge of how to restore shortleaf pine–hardwood mixtures is limited throughout shortleaf pine’s large native range. The objectives of this study were to compare planted shortleaf pine and natural hardwood regeneration survival, growth, and composition following various site preparation and early release treatments. Cluster planting and partial timber harvesting were used to reintroduce shortleaf pine and create two-aged stands in the Cumberland Mountains of Tennessee, USA. Results indicated that shortleaf pine survival, basal diameter, and height growth did not differ following four growing seasons among treatments. Natural regeneration stem densities and heights within shortleaf pine clusters did not differ significantly by treatment. Natural regeneration stem densities differed by species group and height class across the site, while the treatment species interaction term was also significant. -
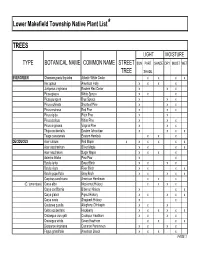
Native Plant List Trees.XLS
Lower Makefield Township Native Plant List* TREES LIGHT MOISTURE TYPE BOTANICAL NAME COMMON NAME STREET SUN PART SHADE DRY MOIST WET TREE SHADE EVERGREEN Chamaecyparis thyoides Atlantic White Cedar x x x x IIex opaca American Holly x x x x Juniperus virginiana Eastern Red Cedar x x x Picea glauca White Spruce x x x Picea pungens Blue Spruce x x x Pinus echinata Shortleaf Pine x x x Pinus resinosa Red Pine x x x Pinus rigida Pitch Pine x x Pinus strobus White Pine x x x Pinus virginiana Virginia Pine x x x Thuja occidentalis Eastern Arborvitae x x x x Tsuga canadensis Eastern Hemlock xx x DECIDUOUS Acer rubrum Red Maple x x x x x x Acer saccharinum Silver Maple x x x x Acer saccharum Sugar Maple x x x x Asimina triloba Paw-Paw x x Betula lenta Sweet Birch x x x x Betula nigra River Birch x x x x Betula populifolia Gray Birch x x x x x Carpinus caroliniana American Hornbeam x x x (C. tomentosa) Carya alba Mockernut Hickory x x x x Carya cordiformis Bitternut Hickory x x x Carya glabra Pignut Hickory x x x x x Carya ovata Shagbark Hickory x x Castanea pumila Allegheny Chinkapin xx x Celtis occidentalis Hackberry x x x x x x Crataegus crus-galli Cockspur Hawthorn x x x x Crataegus viridis Green Hawthorn x x x x Diospyros virginiana Common Persimmon x x x x Fagus grandifolia American Beech x x x x PAGE 1 Exhibit 1 TREES (cont'd) LIGHT MOISTURE TYPE BOTANICAL NAME COMMON NAME STREET SUN PART SHADE DRY MOIST WET TREE SHADE DECIDUOUS (cont'd) Fraxinus americana White Ash x x x x Fraxinus pennsylvanica Green Ash x x x x x Gleditsia triacanthos v. -

Screening Pinus Taeda (Loblolly Pine) Families for Physical and Mechanical Properties Using Vibrational Spectroscopy
SCREENING PINUS TAEDA (LOBLOLLY PINE) FAMILIES FOR PHYSICAL AND MECHANICAL PROPERTIES USING VIBRATIONAL SPECTROSCOPY Gifty E. Acquah, Brian K. Via, Lori G. Eckhardt, and Oladiran O. Fasina1 Abstract—In a bid to control the loblolly pine decline complex, stakeholders are using the selection and deployment of genetically superior families that are disease tolerant. It is vital that we do not compromise other important properties while breeding for disease tolerance. In this preliminary study, near infrared spectroscopy was utilized in conjunction with data collected via conventional methods to develop partial least squares regression models that were used to rapidly predict the basic density, stiffness and ultimate strength of 14 genetically superior loblolly pine families that have been selectively bred to be disease tolerant. Calibration and independent validation data were from southern pines acquired from a commercial sawmill. Seven or eight latent variables were used in model development. The coefficients of determination of the predictive models were more promising for MOE (0.58) and MOR (0.42). INTRODUCTION product. Furthermore, it is essential that strength is not Pinus taeda (loblolly pine) is the most economically compromised else mortality due to reasons other than important tree species in the USA. With 30 million acres forest disease such as wind failure could occur. in plantations in the southern US alone, it provides 110,000 jobs and contributes approximately $30 billion MATERIALS to the economy of this region. However, over the Fourteen year old loblolly pine trees were harvested past 50 years, reduced growth, decline and eventual from two forest sites. Seven families were harvested mortality have been associated with loblolly pine trees from Yulee FL and seven from Waycross GA in February (Eckhardt and others 2010). -

Pinus Echinata, Shortleaf Pine
PinusPinus echinataechinata shortleafshortleaf pinepine by Dr. Kim D. Coder, Professor of Tree Biology & Health Care Warnell School of Forestry & Natural Resources, University of Georgia One of the most widespread pines of the Eastern United Sates is Pinus echinata, shortleaf pine. Shortleaf pine was identified and named in 1768. The scientific name means a “prickly pine cone tree.” Other common names for shortleaf pine include shortstraw pine, yellow pine, Southern yellow pine, shortleaf yellow pine, Arkansas soft pine, Arkansas pine, and old field pine. Among all the Southern yellow pines it has the greatest range and is most tolerant of a variety of sites. Shortleaf pine grows Southeast of a line between New York and Texas. It is widespread in Georgia except for coastal coun- ties. Note the Georgia range map figure. Pinus echinata is found growing in many mixtures with other pines and hardwoods. It tends to grow on medium to dry, well-drained, infertile sites, as compared with loblolly pine (Pinus taeda). It grows quickly in deep, well-drained areas of floodplains, but cannot tolerate high pH and high calcium concentrations. Compared with other Southern yellow pines, shortleaf is less demanding of soil oxygen content and essential element availability. It grows in Hardiness Zone 6a - 8b and Heat Zone 6-9. The lowest number of Hardiness Zone tends to delineate the Northern range limit and the largest Heat Zone number tends to define the South- ern edge of the range. This native Georgia pine grows in Coder Tree Grow Zone (CTGZ) A-D (a mul- tiple climatic attribute based map), and in the temperature and precipitation cluster based Coder Tree Planting Zone 1-6.