Androgenic to Estrogenic Switch Due to Epigenetic Silencing of Steroid 5-Α Reductase 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Association Between Two Polymorphisms in the SRD5A2 Gene and Serum Androgen Concentrations in British Men1
578 Vol. 12, 578–581, June 2003 Cancer Epidemiology, Biomarkers & Prevention Short Communication Association between Two Polymorphisms in the SRD5A2 Gene and Serum Androgen Concentrations in British Men1 Naomi E. Allen,2 Juergen K. V. Reichardt, are needed to additionally clarify the role of the A49T Hannah Nguyen, and Timothy J. Key polymorphism in androgen metabolism. Cancer Research United Kingdom Epidemiology Unit, University of Oxford, Gibson Building, Radcliffe Infirmary, Oxford, OX2 6HE, United Kingdom Introduction [N. E. A., T. J. K.], and Institute for Genetic Medicine, Departments of Biochemistry and Molecular Biology and Preventive Medicine, University of It is well established that the steroid 5 ␣-reductase type II Southern California School of Medicine, University of Southern California, enzyme, which irreversibly converts testosterone to its more Los Angeles, California 90089-9075 [J. K. V. R., H. N.] potent metabolite DHT3 in prostatic cells, is required for the normal growth and development of the prostate gland (1). A meta-analysis of prospective studies suggests that the serum Abstract concentration of A-diol-g, a serum marker of the abundance of Androgens are essential for the growth of the prostate 5 ␣-reductase type II and intraprostatic DHT, is slightly higher gland and have been implicated in the development of in men who subsequently develop prostate cancer compared prostate cancer. Little is known about the determinants with healthy individuals (2). The circulating A-diol-g concen- of androgen levels in men, although observed ethnic tration is believed to be a more accurate serum marker of 5 differences suggest they may have a genetic basis. -

Type of the Paper (Article
Supplementary Material A Proteomics Study on the Mechanism of Nutmeg-induced Hepatotoxicity Wei Xia 1, †, Zhipeng Cao 1, †, Xiaoyu Zhang 1 and Lina Gao 1,* 1 School of Forensic Medicine, China Medical University, Shenyang 110122, P. R. China; lessen- [email protected] (W.X.); [email protected] (Z.C.); [email protected] (X.Z.) † The authors contributed equally to this work. * Correspondence: [email protected] Figure S1. Table S1. Peptide fraction separation liquid chromatography elution gradient table. Time (min) Flow rate (mL/min) Mobile phase A (%) Mobile phase B (%) 0 1 97 3 10 1 95 5 30 1 80 20 48 1 60 40 50 1 50 50 53 1 30 70 54 1 0 100 1 Table 2. Liquid chromatography elution gradient table. Time (min) Flow rate (nL/min) Mobile phase A (%) Mobile phase B (%) 0 600 94 6 2 600 83 17 82 600 60 40 84 600 50 50 85 600 45 55 90 600 0 100 Table S3. The analysis parameter of Proteome Discoverer 2.2. Item Value Type of Quantification Reporter Quantification (TMT) Enzyme Trypsin Max.Missed Cleavage Sites 2 Precursor Mass Tolerance 10 ppm Fragment Mass Tolerance 0.02 Da Dynamic Modification Oxidation/+15.995 Da (M) and TMT /+229.163 Da (K,Y) N-Terminal Modification Acetyl/+42.011 Da (N-Terminal) and TMT /+229.163 Da (N-Terminal) Static Modification Carbamidomethyl/+57.021 Da (C) 2 Table S4. The DEPs between the low-dose group and the control group. Protein Gene Fold Change P value Trend mRNA H2-K1 0.380 0.010 down Glutamine synthetase 0.426 0.022 down Annexin Anxa6 0.447 0.032 down mRNA H2-D1 0.467 0.002 down Ribokinase Rbks 0.487 0.000 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2
biology Article Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2 Chiranan Khantham 1 , Wipawadee Yooin 1,2 , Korawan Sringarm 2,3 , Sarana Rose Sommano 2,4 , Supat Jiranusornkul 1, Francisco David Carmona 5,6 , Wutigri Nimlamool 7 , Pensak Jantrawut 1,2, Pornchai Rachtanapun 8,9 and Warintorn Ruksiriwanich 1,2,8,* 1 Department of Pharmaceutical Sciences, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (C.K.); [email protected] (W.Y.); [email protected] (S.J.); [email protected] (P.J.) 2 Cluster of Research and Development of Pharmaceutical and Natural Products Innovation for Human or Animal, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (K.S.); [email protected] (S.R.S.) 3 Department of Animal and Aquatic Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 4 Plant Bioactive Compound Laboratory (BAC), Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 5 Departamento de Genética e Instituto de Biotecnología, Universidad de Granada, 18071 Granada, Spain; [email protected] 6 Instituto de Investigación Biosanitaria ibs.GRANADA, 18014 Granada, Spain 7 Citation: Khantham, C.; Yooin, W.; Department of Pharmacology, Faculty of Medicine, Chiang Mai University, Chiang Mai 50200, Thailand; Sringarm, K.; Sommano, S.R.; [email protected] 8 Cluster of Agro Bio-Circular-Green Industry, Faculty of Agro-Industry, Chiang Mai University, Jiranusornkul, S.; Carmona, F.D.; Chiang Mai 50100, Thailand; [email protected] Nimlamool, W.; Jantrawut, P.; 9 School of Agro-Industry, Faculty of Agro-Industry, Chiang Mai University, Chiang Mai 50100, Thailand Rachtanapun, P.; Ruksiriwanich, W. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Crystal Structure of Steroid Reductase SRD5A Reveals Conserved Steroid Reduction Mechanism
ARTICLE https://doi.org/10.1038/s41467-020-20675-2 OPEN Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism Yufei Han 1,9, Qian Zhuang2,9, Bo Sun3,9, Wenping Lv 4,9, Sheng Wang5,9, Qingjie Xiao6, Bin Pang1, ✉ Youli Zhou1, Fuxing Wang1, Pengliang Chi6, Qisheng Wang3, Zhen Li7, Lizhe Zhu 4, Fuping Li8, Dong Deng6 , ✉ ✉ ✉ Ying-Chih Chiang 1 , Zhenfei Li 2 & Ruobing Ren 1 Steroid hormones are essential in stress response, immune system regulation, and repro- 1234567890():,; duction in mammals. Steroids with 3-oxo-Δ4 structure, such as testosterone or progesterone, are catalyzed by steroid 5α-reductases (SRD5As) to generate their corresponding 3-oxo-5α steroids, which are essential for multiple physiological and pathological processes. SRD5A2 is already a target of clinically relevant drugs. However, the detailed mechanism of SRD5A- mediated reduction remains elusive. Here we report the crystal structure of PbSRD5A from Proteobacteria bacterium, a homolog of both SRD5A1 and SRD5A2, in complex with the cofactor NADPH at 2.0 Å resolution. PbSRD5A exists as a monomer comprised of seven transmembrane segments (TMs). The TM1-4 enclose a hydrophobic substrate binding cavity, whereas TM5-7 coordinate cofactor NADPH through extensive hydrogen bonds network. Homology-based structural models of HsSRD5A1 and -2, together with biochemical char- acterization, define the substrate binding pocket of SRD5As, explain the properties of disease-related mutants and provide an important framework for further understanding of the mechanism of NADPH mediated steroids 3-oxo-Δ4 reduction. Based on these analyses, the design of therapeutic molecules targeting SRD5As with improved specificity and therapeutic efficacy would be possible. -

Bioactivity of Curcumin on the Cytochrome P450 Enzymes of the Steroidogenic Pathway
International Journal of Molecular Sciences Article Bioactivity of Curcumin on the Cytochrome P450 Enzymes of the Steroidogenic Pathway Patricia Rodríguez Castaño 1,2, Shaheena Parween 1,2 and Amit V Pandey 1,2,* 1 Pediatric Endocrinology, Diabetology, and Metabolism, University Children’s Hospital Bern, 3010 Bern, Switzerland; [email protected] (P.R.C.); [email protected] (S.P.) 2 Department of Biomedical Research, University of Bern, 3010 Bern, Switzerland * Correspondence: [email protected]; Tel.: +41-31-632-9637 Received: 5 September 2019; Accepted: 16 September 2019; Published: 17 September 2019 Abstract: Turmeric, a popular ingredient in the cuisine of many Asian countries, comes from the roots of the Curcuma longa and is known for its use in Chinese and Ayurvedic medicine. Turmeric is rich in curcuminoids, including curcumin, demethoxycurcumin, and bisdemethoxycurcumin. Curcuminoids have potent wound healing, anti-inflammatory, and anti-carcinogenic activities. While curcuminoids have been studied for many years, not much is known about their effects on steroid metabolism. Since many anti-cancer drugs target enzymes from the steroidogenic pathway, we tested the effect of curcuminoids on cytochrome P450 CYP17A1, CYP21A2, and CYP19A1 enzyme activities. When using 10 µg/mL of curcuminoids, both the 17α-hydroxylase as well as 17,20 lyase activities of CYP17A1 were reduced significantly. On the other hand, only a mild reduction in CYP21A2 activity was observed. Furthermore, CYP19A1 activity was also reduced up to ~20% of control when using 1–100 µg/mL of curcuminoids in a dose-dependent manner. Molecular docking studies confirmed that curcumin could dock onto the active sites of CYP17A1, CYP19A1, as well as CYP21A2. -

Original Article Genetic Polymorphisms in the Androgen Metabolism Pathway and Risk of Prostate Cancer in Low Incidence Malaysian Ethnic Groups
Int J Clin Exp Med 2015;8(10):19232-19240 www.ijcem.com /ISSN:1940-5901/IJCEM0011564 Original Article Genetic polymorphisms in the androgen metabolism pathway and risk of prostate cancer in low incidence Malaysian ethnic groups Prevathe Poniah1, Zahurin Mohamed2, Yamunah Devi Apalasamy2, Shamsul Mohd Zain2, Shanggar Kuppusamy1, Azad HA Razack1 Departments of 1Surgery, 2Pharmacology, Faculty of Medicine, University of Malaya, Kuala Lumpur 50603, Malay- sia Received June 17, 2015; Accepted September 22, 2015; Epub October 15, 2015; Published October 30, 2015 Abstract: Androgens are involved in prostate cancer (PCa) cell growth. Genes involved in androgen metabolism mediate key steps in sex steroid metabolism. This study attempted to investigate whether single nucleotide poly- morphisms (SNPs) in the androgen metabolism pathway are associated with PCa risk in low incidence Asian ethnic groups. We genotyped 172 Malaysian subjects for cytochrome P450 family 17 (CYP17A1), steroid-5-alpha-reduc- tase, polypeptide 1 and 2 (SRD5A1 and SRD5A2), and insulin-like growth factor 1 (IGF-1) genes of the androgen metabolism pathway and assessed the testosterone, dihydrotestosterone and IGF-1 levels. SNPs in the CYP17A1, SRD5A1, SRD5A2, and IGF-1 genes were genotyped using real-time polymerase chain reaction. Although we did not find significant association between SNPs analysed in this study with PCa risk, we observed however, signifi- cant association between androgen levels and the IGF-1 and several SNPs. Men carrying the GG genotype for SNP rs1004467 (CYP17A1) had significantly elevated testosterone (P = 0.012) and dihydrotestosterone (DHT) levels (P = 0.024) as compared to carriers of the A allele. The rs518673 of the SRD5A1 was associated with prostate spe- cific antigen (PSA) levels. -

ERG/AKR1C3/AR Constitutes a Feed-Forward Loop for AR Signaling in Prostate Cancer Cells Katelyn Powell1, Louie Semaan1, M
Published OnlineFirst March 9, 2015; DOI: 10.1158/1078-0432.CCR-14-2352 Biology of Human Tumors Clinical Cancer Research ERG/AKR1C3/AR Constitutes a Feed-Forward Loop for AR Signaling in Prostate Cancer Cells Katelyn Powell1, Louie Semaan1, M. Katie Conley-LaComb1, Irfan Asangani2, Yi-Mi Wu2, Kevin B. Ginsburg1, Julia Williams3, Jeremy A. Squire4, Krishna R. Maddipati5, Michael L. Cher1,5,6, and Sreenivasa R. Chinni1,5,6 Abstract Purpose: Intratumoral androgen synthesis in prostate cancer Results: We found that ERG regulated the expression of the ABE contributes to the development of castration-resistant prostate AKR1C3 in prostate cancer cells via direct binding to the AKR1C3 cancer (CRPC). Several enzymes responsible for androgen bio- gene. Knockdown of ERG resulted in reduced AKR1C3 expression, synthesis have been shown to be overexpressed in CRPC, thus which caused a reduction in both DHT synthesis and PSA expres- contributing to CRPC in a castrated environment. The TMPRSS2– sion in VCaP prostate cancer cells treated with 5a-androstane- ERG transcription factor has been shown to be present in primary dione (5a-Adione), a DHT precursor metabolite. Immunohisto- prostate cancer tumors as well as CRPC tumors. We hypothesize chemical staining revealed that ERG was coexpressed with that TMPRSS2–ERG fusions regulate androgen biosynthetic AKR1C3 in prostate cancer tissue samples. enzyme (ABE) gene expression and the production of androgens, Conclusions: These data suggest that AKR1C3 catalyzes the which contributes to the development of CRPC. biochemical reduction of 5a-Adione to DHT in prostate cancer Experimental design: We used a panel of assays, including cells, and that ERG regulates this step through upregulation of lentivirus transduction, gene expression, chromatin immunopre- AKR1C3 expression. -

Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models
cancers Article Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models Subrata Deb 1 , Steven Pham 2, Dong-Sheng Ming 2, Mei Yieng Chin 2, Hans Adomat 2, Antonio Hurtado-Coll 2, Martin E. Gleave 2,3 and Emma S. Tomlinson Guns 2,3,* 1 Department of Pharmaceutical Sciences, College of Pharmacy, Larkin University, Miami, FL 33169, USA; [email protected] 2 The Vancouver Prostate Centre at Vancouver General Hospital, 2660 Oak Street, Vancouver, BC V6H 3Z6, Canada; [email protected] (S.P.); [email protected] (D.-S.M.); [email protected] (M.Y.C.); [email protected] (H.A.); [email protected] (A.H.-C.); [email protected] (M.E.G.) 3 Department of Urologic Sciences, Faculty of Medicine, University of British Columbia, Vancouver, BC V5Z 1M9, Canada * Correspondence: [email protected]; Tel.: +1-604-875-4818 Received: 14 August 2018; Accepted: 17 September 2018; Published: 20 September 2018 Abstract: Castration-resistant prostate tumors acquire the independent capacity to generate androgens by upregulating steroidogenic enzymes or using steroid precursors produced by the adrenal glands for continued growth and sustainability. The formation of steroids was measured by liquid chromatography-mass spectrometry in LNCaP and 22Rv1 prostate cancer cells, and in human prostate tissues, following incubation with steroid precursors (22-OH-cholesterol, pregnenolone, 17-OH-pregnenolone, progesterone, 17-OH-progesterone). Pregnenolone, progesterone, 17-OH-pregnenolone, and 17-OH-progesterone increased C21 steroid (5-pregnan-3,20-dione, 5-pregnan-3,17-diol-20-one, 5-pregnan-3-ol-20-one) formation in the backdoor pathway, and demonstrated a trend of stimulating dihydroepiandrosterone or its precursors in the backdoor pathway in LNCaP and 22Rv1 cells. -
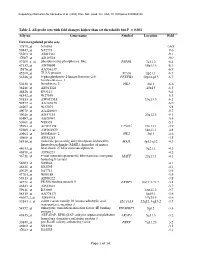
Table 2. All Probe Sets with Fold Changes Higher Than Set Thresholds but P > 0.001 Affy No
Supporting information for Sanoudou et al. (2003) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0330960100 Table 2. All probe sets with fold changes higher than set thresholds but P > 0.001 Affy no. Gene name Symbol Location Fold Down-regulated probe sets 47879_at N46863 -16.9 59447_at N52773 -9.6 55503_at AI085361 -9.1 47087_at AI310524 -9.0 37209_g_at phosphoserine phosphatase-like PSPHL 7q11.2 -8.4 47137_at AI479899 19p13.3 -8.1 45876_at AA536137 -6.9 45260_at TU3A protein TU3A 3p21.1 -6.7 56246_at 6-phosphofructo-2-kinase/fructose-2,6- PFKFB3 10p14-p15 -6.7 bisphosphatase 3 50230_at hexokinase 2 HK2 2p13 -6.6 38248_at AB011124 20p13 -6.5 44426_at R93141 -6.4 48542_at W27559 -6.3 53813_at AW051518 13q13.3 -6.1 59577_at AA243670 -6.0 46907_at W37075 -5.8 49078_at AA424983 -5.7 49026_at AI357153 20p12.3 -5.4 53487_at AI670947 -5.4 50161_at N39328 -5.1 35994_at AC002398 F25965 19q13.1 -4.9 52285_f_at AW002970 18p11.1 -4.8 40964_at hexokinase 2 HK2 2p13 -4.6 49806_at AI932283 -4.5 58918_at molecule possessing ankyrin repeats induced by MAIL 3p12-q12 -4.3 lipopolysaccharide (MAIL), homolog of mouse 44633_at heat shock 27 kDa associated protein 3q21.1 -4.3 46858_at AI796221 -4.2 36711_at v-maf musculoaponeurotic fibrosarcoma oncogene MAFF 22q13.1 -4.1 homolog F (avian) 54683_at N49844 -4.1 46621_at N32595 -4.1 49629_at N47713 -3.9 47703_at W89189 -3.9 59313_at AI598222 -3.8 34721_at FK506 binding protein 5 FKBP5 6p21.3-21.2 -3.8 46843_at AI632621 -3.7 59611_at R53069 16p11.2 -3.7 58315_at AA778171 3p25.1 -3.6 46607_f_at AI885018 17q25.3 -3.6 33143_s_at solute carrier family 16 (monocarboxylic acid SLC16A3 22q12.3-q13.2 -3.5 transporters), member 3 54152_at eukaryotic translation initiation factor 4E binding EIF4EBP1 8p12 -3.4 protein 1 43935_at ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin ARAP3 5q31.3 -3.2 homology domains-containing protein 3 33849_at pre-B-cell colony-enhancing factor PBEF 7q11.23 -3.2 46902_at N92294 -3.2 47023_at N25555 -3.1 Page 1 of 14 Supporting information for Sanoudou et al. -

Metabolic Enzyme Expression Highlights a Key Role for MTHFD2 and the Mitochondrial Folate Pathway in Cancer
ARTICLE Received 1 Nov 2013 | Accepted 17 Dec 2013 | Published 23 Jan 2014 DOI: 10.1038/ncomms4128 Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer Roland Nilsson1,2,*, Mohit Jain3,4,5,6,*,w, Nikhil Madhusudhan3,4,5, Nina Gustafsson Sheppard1,2, Laura Strittmatter3,4,5, Caroline Kampf7, Jenny Huang8, Anna Asplund7 & Vamsi K. Mootha3,4,5 Metabolic remodeling is now widely regarded as a hallmark of cancer, but it is not clear whether individual metabolic strategies are frequently exploited by many tumours. Here we compare messenger RNA profiles of 1,454 metabolic enzymes across 1,981 tumours spanning 19 cancer types to identify enzymes that are consistently differentially expressed. Our meta- analysis recovers established targets of some of the most widely used chemotherapeutics, including dihydrofolate reductase, thymidylate synthase and ribonucleotide reductase, while also spotlighting new enzymes, such as the mitochondrial proline biosynthetic enzyme PYCR1. The highest scoring pathway is mitochondrial one-carbon metabolism and is centred on MTHFD2. MTHFD2 RNA and protein are markedly elevated in many cancers and correlated with poor survival in breast cancer. MTHFD2 is expressed in the developing embryo, but is absent in most healthy adult tissues, even those that are proliferating. Our study highlights the importance of mitochondrial compartmentalization of one-carbon metabolism in cancer and raises important therapeutic hypotheses. 1 Unit of Computational Medicine, Department of Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 2 Center for Molecular Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 3 Broad Institute, Cambridge, Massachusetts 02142, USA. 4 Department of Systems Biology, Harvard Medical School, Boston, Massachusetts 02115, USA.