Subunitspecific Inhibition of Acid Sensing Ion Channels by Stomatinlike Protein 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptomic Analysis of Native Versus Cultured Human and Mouse Dorsal Root Ganglia Focused on Pharmacological Targets Short
bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Transcriptomic analysis of native versus cultured human and mouse dorsal root ganglia focused on pharmacological targets Short title: Comparative transcriptomics of acutely dissected versus cultured DRGs Andi Wangzhou1, Lisa A. McIlvried2, Candler Paige1, Paulino Barragan-Iglesias1, Carolyn A. Guzman1, Gregory Dussor1, Pradipta R. Ray1,#, Robert W. Gereau IV2, # and Theodore J. Price1, # 1The University of Texas at Dallas, School of Behavioral and Brain Sciences and Center for Advanced Pain Studies, 800 W Campbell Rd. Richardson, TX, 75080, USA 2Washington University Pain Center and Department of Anesthesiology, Washington University School of Medicine # corresponding authors [email protected], [email protected] and [email protected] Funding: NIH grants T32DA007261 (LM); NS065926 and NS102161 (TJP); NS106953 and NS042595 (RWG). The authors declare no conflicts of interest Author Contributions Conceived of the Project: PRR, RWG IV and TJP Performed Experiments: AW, LAM, CP, PB-I Supervised Experiments: GD, RWG IV, TJP Analyzed Data: AW, LAM, CP, CAG, PRR Supervised Bioinformatics Analysis: PRR Drew Figures: AW, PRR Wrote and Edited Manuscript: AW, LAM, CP, GD, PRR, RWG IV, TJP All authors approved the final version of the manuscript. 1 bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

The Chondrocyte Channelome: a Novel Ion Channel Candidate in the Pathogenesis of Pectus Deformities
Old Dominion University ODU Digital Commons Biological Sciences Theses & Dissertations Biological Sciences Summer 2017 The Chondrocyte Channelome: A Novel Ion Channel Candidate in the Pathogenesis of Pectus Deformities Anthony J. Asmar Old Dominion University, [email protected] Follow this and additional works at: https://digitalcommons.odu.edu/biology_etds Part of the Biology Commons, Molecular Biology Commons, and the Physiology Commons Recommended Citation Asmar, Anthony J.. "The Chondrocyte Channelome: A Novel Ion Channel Candidate in the Pathogenesis of Pectus Deformities" (2017). Doctor of Philosophy (PhD), Dissertation, Biological Sciences, Old Dominion University, DOI: 10.25777/pyha-7838 https://digitalcommons.odu.edu/biology_etds/19 This Dissertation is brought to you for free and open access by the Biological Sciences at ODU Digital Commons. It has been accepted for inclusion in Biological Sciences Theses & Dissertations by an authorized administrator of ODU Digital Commons. For more information, please contact [email protected]. THE CHONDROCYTE CHANNELOME: A NOVEL ION CHANNEL CANDIDATE IN THE PATHOGENESIS OF PECTUS DEFORMITIES by Anthony J. Asmar B.S. Biology May 2010, Virginia Polytechnic Institute M.S. Biology May 2013, Old Dominion University A Dissertation Submitted to the Faculty of Old Dominion University in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY BIOMEDICAL SCIENCES OLD DOMINION UNIVERSITY August 2017 Approved by: Christopher Osgood (Co-Director) Michael Stacey (Co-Director) Lesley Greene (Member) Andrei Pakhomov (Member) Jing He (Member) ABSTRACT THE CHONDROCYTE CHANNELOME: A NOVEL ION CHANNEL CANDIDATE IN THE PATHOGENESIS OF PECTUS DEFORMITIES Anthony J. Asmar Old Dominion University, 2017 Co-Directors: Dr. Christopher Osgood Dr. Michael Stacey Costal cartilage is a type of rod-like hyaline cartilage connecting the ribs to the sternum. -

Transcriptomic Profiling of Ca Transport Systems During
cells Article Transcriptomic Profiling of Ca2+ Transport Systems during the Formation of the Cerebral Cortex in Mice Alexandre Bouron Genetics and Chemogenomics Lab, Université Grenoble Alpes, CNRS, CEA, INSERM, Bâtiment C3, 17 rue des Martyrs, 38054 Grenoble, France; [email protected] Received: 29 June 2020; Accepted: 24 July 2020; Published: 29 July 2020 Abstract: Cytosolic calcium (Ca2+) transients control key neural processes, including neurogenesis, migration, the polarization and growth of neurons, and the establishment and maintenance of synaptic connections. They are thus involved in the development and formation of the neural system. In this study, a publicly available whole transcriptome sequencing (RNA-Seq) dataset was used to examine the expression of genes coding for putative plasma membrane and organellar Ca2+-transporting proteins (channels, pumps, exchangers, and transporters) during the formation of the cerebral cortex in mice. Four ages were considered: embryonic days 11 (E11), 13 (E13), and 17 (E17), and post-natal day 1 (PN1). This transcriptomic profiling was also combined with live-cell Ca2+ imaging recordings to assess the presence of functional Ca2+ transport systems in E13 neurons. The most important Ca2+ routes of the cortical wall at the onset of corticogenesis (E11–E13) were TACAN, GluK5, nAChR β2, Cav3.1, Orai3, transient receptor potential cation channel subfamily M member 7 (TRPM7) non-mitochondrial Na+/Ca2+ exchanger 2 (NCX2), and the connexins CX43/CX45/CX37. Hence, transient receptor potential cation channel mucolipin subfamily member 1 (TRPML1), transmembrane protein 165 (TMEM165), and Ca2+ “leak” channels are prominent intracellular Ca2+ pathways. The Ca2+ pumps sarco/endoplasmic reticulum Ca2+ ATPase 2 (SERCA2) and plasma membrane Ca2+ ATPase 1 (PMCA1) control the resting basal Ca2+ levels. -

Mechanosensitive Channels: Multiplicity of Families and Gating Paradigms
R EVIEW Mechanosensitive Channels: Multiplicity of Families and Gating Paradigms Sergei Sukharev1* and David P. Corey2* (Published 10 February 2004) Mechanosensitive ion channels are the primary and comprise just a few GTP-binding protein (G protein)–cou- transducers that convert mechanical force into an pled receptor second-messenger pathways (1–3). Given these electrical or chemical signal in hearing, touch, and examples, should we be looking for universal mechanotransduc- other mechanical senses. Unlike vision, olfaction, tion mechanisms within these various systems and presume that and some types of taste, which all use similar kinds the primary receiver elements are essentially the same, but are of primary heterotrimeric GTP-binding protein–cou- tuned differently depending on the setting? If not, are there dif- pled receptors, mechanosensation relies on diverse ferent types of mechanoreceptors that use similar biophysical types of transducer molecules. Unrelated types of principles but dissimilar molecular mechanisms, which arose channels can be used for the perception of various independently many times in the course of evolution? The ex- mechanical stimuli, not only in distant groups of or- amples discussed below suggest that the latter scenario is more ganisms, but also in separate locations of the same likely. Although incomplete, the available molecular data sug- organism. The extreme sensitivity of the transduction gest a multiplicity of structural designs and mechanisms of mechanism in the auditory system, which relies on mechanoreceptors not only across the tree of life, but also with- an elaborate structure of rigid cilia, filamentous in single organisms. This diversity implies that careful analysis links, and molecular motors to focus force on trans- of individual systems will be required before generalizations duction channels, contrasts with that of the bacterial can be made. -

Genetic Mapping of ASIC4 and Contrasting Phenotype to Asic1a in Modulating Innate Fear and Anxiety
European Journal of Neuroscience, Vol. 41, pp. 1553–1568, 2015 doi:10.1111/ejn.12905 BEHAVIORAL NEUROSCIENCE Genetic mapping of ASIC4 and contrasting phenotype to ASIC1a in modulating innate fear and anxiety Shing-Hong Lin,1,2 Ya-Chih Chien,2 Wei-Wei Chiang,3 Yan-Zhen Liu,3 Cheng-Chang Lien4 and Chih-Cheng Chen1,2,3 1Graduate institute of Life Sciences, National Defense Medical Center, Taipei, Taiwan 2Institute of Biomedical Sciences, Academia Sinica, Taipei 115, Taiwan 3Taiwan Mouse Clinic-National Comprehensive Mouse Phenotyping and Drug Testing Center, Academia Sinica, Taipei, Taiwan 4Institute of Neuroscience, National Yang-Ming University, Taipei, Taiwan Keywords: ASIC, behavioral phenotyping, Cre, interneuron, knockout mice Abstract Although ASIC4 is a member of the acid-sensing ion channel (ASIC) family, we have limited knowledge of its expression and physiological function in vivo. To trace the expression of this ion channel, we generated the ASIC4-knockout/CreERT2-knockin (Asic4CreERT2) mouse line. After tamoxifen induction in the Asic4CreERT2::CAG-STOPfloxed-Td-tomato double transgenic mice, we mapped the expression of ASIC4 at the cellular level in the central nervous system (CNS). ASIC4 was expressed in many brain regions, including the olfactory bulb, cerebral cortex, striatum, hippocampus, amygdala, thalamus, hypothalamus, brain stem, cer- ebellum, spinal cord and pituitary gland. Colocalisation studies further revealed that ASIC4 was expressed mainly in three types of cells in the CNS: (i) calretinin (CR)-positive and/or vasoactive intestine peptide (VIP)-positive interneurons; (ii) neural/glial anti- gen 2 (NG2)-positive glia, also known as oligodendrocyte precursor cells; and (iii) cerebellar granule cells. To probe the possible role of ASIC4, we hypothesised that ASIC4 could modulate the membrane expression of ASIC1a and thus ASIC1a signaling in vivo. -

Multiple Modulation of Acid-Sensing Ion Channel 1A by the Alkaloid Daurisoline
biomolecules Article Multiple Modulation of Acid-Sensing Ion Channel 1a by the Alkaloid Daurisoline Dmitry I. Osmakov 1,2,* , Sergey G. Koshelev 1, Ekaterina N. Lyukmanova 1, Mikhail A. Shulepko 1, Yaroslav A. Andreev 1,2, Peter Illes 3 and Sergey A. Kozlov 1 1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, 117997 Moscow, Russia 2 Institute of Molecular Medicine, Sechenov First Moscow State Medical University, Trubetskaya str. 8, bld. 2, 119991 Moscow, Russia 3 Rudolf-Boehm-Institut für Pharmakologie und Toxikologie, University of Leipzig, 04107 Leipzig, Germany * Correspondence: [email protected] Received: 26 June 2019; Accepted: 1 August 2019; Published: 2 August 2019 Abstract: Acid-sensing ion channels (ASICs) are proton-gated sodium-selective channels that are expressed in the peripheral and central nervous systems. ASIC1a is one of the most intensively studied isoforms due to its importance and wide representation in organisms, but it is still largely unexplored as a target for therapy. In this study, we demonstrated response of the ASIC1a to acidification in the presence of the daurisoline (DAU) ligand. DAU alone did not activate the channel, but in combination with protons, it produced the second peak component of the ASIC1a current. This second peak differs from the sustained component (which is induced by RF-amide peptides), as the second (DAU-induced) peak is completely desensitized, with the same kinetics as the main peak. The co-application of DAU and mambalgin-2 indicated that their binding sites do not overlap. Additionally,we found an asymmetry in the pH activation curve of the channel, which was well-described by a mathematical model based on the multiplied probabilities of protons binding with a pool of high-cooperative sites and a single proton binding with a non-cooperative site. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -
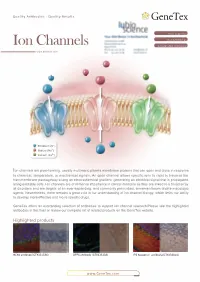
Ion Channels Accelerated D1scovery
Quality Antibodies · Quality Results $-GeneTex Your Expertise Our Antibod1es Ion Channels Accelerated D1scovery --------- www.genetex.com Potassium (K+ ) Sodium (Na+ ) Calcium (Ca2+ ) Ion channels are pore-forming, usually multimeric plasma membrane proteins that can open and close in response to chemical, temperature, or mechanical signals. An open channel allows specific ions to rapid ly traverse the transmembrane passageway a long an electrochemical gradient, generating an electrical signal that is propagated along excitable cells. Ion channels are of immense importance in clinical medicine as they are linked to a broad array of disorders and are targets of an ever-expanding, and commonly prescribed, armamentarium of pharmacologic agents. Nevertheless, there remains a great void in our understanding of ion channel biology, which limits our ability to develop more effective andmo re specific drugs. GeneTex offers an outstanding selection of antibodies to support ion channel research.Please see the highlighted antibodies in this flyer or review our complete list of related products on the GeneTex website. Highlighted products HCNl antibody (GTX131334} DPP6 antibody (GTX133338} IP3 Receptor I antibody (GTX133104} (...___ ___w _w _w_ ._G_e_n_e_T_e_x_._c_o_m_ _____,) ` a n a 匱 。 'ot I: . ·""·,, " . 鼴 丶•' · ,' , `> ,,, 丶 B -- ,:: ·-- o .i :' T. 、 冨 ,. \\.◄ .•}' r<• .. , -·•Jt.<_,,,, . ◄ · ◄ / .Jj -~~ 盆;i . '. ., CACNB4 (GTX100202) VGluTl antibody (GTX133148) P2X7一 antibody (GTX104288) Cavl.2 antibody (GTX54754) Cav2.1 antibody (GTX54753) -
A Variant of ASIC2 Mediates Sodium Retention in Nephrotic Syndrome
A variant of ASIC2 mediates sodium retention in nephrotic syndrome Marc Fila, … , Gilles Crambert, Alain Doucet JCI Insight. 2021. https://doi.org/10.1172/jci.insight.148588. Research In-Press Preview Nephrology Graphical abstract Find the latest version: https://jci.me/148588/pdf 1 A variant of ASIC2 mediates sodium retention in nephrotic syndrome 1,2Marc Fila *, 1,2Ali Sassi*, 1,2Gaëlle Brideau, 1,2Lydie Cheval, 1,2 Luciana Morla, 1,2Pascal Houillier, 1,2Christine Walter, 1,2Michel Gennaoui, 1,2Laure Collignon L, 1,2Mathilde Keck, 1,2Gabrielle Planelles, 1,2Naziha Bakouh, 3Michel Peuchmaur, 4Georges Deschênes, 5Ignacio Anegon, 5Séverine Remy, 6Bruno Vogt, 1,2 Gilles Crambert#, 1,2Alain Doucet 1Centre de Recherche des Cordeliers, Sorbonne Universités, INSERM, Université de Paris, Laboratoire de Physiologie Rénale et Tubulopathies, F-75006, Paris, France 2CNRS, ERL8228, F-75006, Paris, France 3Cytology and Pathology DePartment, Robert Debré HosPital, F-75019, Paris, France 4Pediatric NePhrology DePartment, Robert Debré HosPital, F-75019, Paris, France 5INSERM UMR 1064, Centre de Recherches en TransPlantation et Immunologie (CRTI), Transgenic Rats ImmunoPhenomic facility, Nantes, France. 6DePartment of NePhrology and HyPertension, InselsPital, Bern University HosPital, CH- 3010, Bern, Switzerland Present address: Marc Fila, Pediatric NePhrology DePartment, HôPital Arnaud de Villeneuve Institut de Génomique Fonctionnelle UMR9023 CNRS U661 INSERM, MontPellier, France; Ali Sassi, DePartment of Cellular Physiology and Metabolism, University -

1 1 2 3 Cell Type-Specific Transcriptomics of Hypothalamic
1 2 3 4 Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to 5 weight-loss 6 7 Fredrick E. Henry1,†, Ken Sugino1,†, Adam Tozer2, Tiago Branco2, Scott M. Sternson1,* 8 9 1Janelia Research Campus, Howard Hughes Medical Institute, 19700 Helix Drive, Ashburn, VA 10 20147, USA. 11 2Division of Neurobiology, Medical Research Council Laboratory of Molecular Biology, 12 Cambridge CB2 0QH, UK 13 14 †Co-first author 15 *Correspondence to: [email protected] 16 Phone: 571-209-4103 17 18 Authors have no competing interests 19 1 20 Abstract 21 Molecular and cellular processes in neurons are critical for sensing and responding to energy 22 deficit states, such as during weight-loss. AGRP neurons are a key hypothalamic population 23 that is activated during energy deficit and increases appetite and weight-gain. Cell type-specific 24 transcriptomics can be used to identify pathways that counteract weight-loss, and here we 25 report high-quality gene expression profiles of AGRP neurons from well-fed and food-deprived 26 young adult mice. For comparison, we also analyzed POMC neurons, an intermingled 27 population that suppresses appetite and body weight. We find that AGRP neurons are 28 considerably more sensitive to energy deficit than POMC neurons. Furthermore, we identify cell 29 type-specific pathways involving endoplasmic reticulum-stress, circadian signaling, ion 30 channels, neuropeptides, and receptors. Combined with methods to validate and manipulate 31 these pathways, this resource greatly expands molecular insight into neuronal regulation of 32 body weight, and may be useful for devising therapeutic strategies for obesity and eating 33 disorders. -

Naked Mole-Rat Acid-Sensing Ion Channel 3 Forms Nonfunctional Homomers, but Functional Heteromers
bioRxiv preprint doi: https://doi.org/10.1101/166272; this version posted November 16, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Naked mole-rat ASIC3 is proton insensitive Naked mole-rat acid-sensing ion channel 3 forms nonfunctional homomers, but functional heteromers Laura-Nadine Schuhmacher1,2, Gerard Callejo1, Shyam Srivats1,3 and Ewan St. John Smith1* 1Department of Pharmacology, University of Cambridge, Tennis Court Road, Cambridge, CB2 1PD, United Kingdom 2Current address: Department of Cell & Developmental Biology, University College London, Gower Street, London, WC1E 6BT, United Kingdom 3Cero Therapeutics, Oyster Point Boulevard, South San Francisco, California 94080, USA Running title: Naked mole-rat ASIC3 is proton insensitive To whom correspondence should be addressed: Ewan St. John Smith, Department of Pharmacology, University of Cambridge, Tennis Court Road, Cambridge, CB2 1PD, United Kingdom, Tel.: +44 1223 334048; Fax: +44 1223 334100; E-mail: [email protected] Keywords: Acid sensing ion channel (ASIC), pain, ion channel, neuron, neurophysiology ABSTRACT underlie the proton insensitivity of nmrASIC3. Acid-sensing ion channels (ASICs) form both However, introducing nmrASIC3 differences into homotrimeric and heterotrimeric ion channels that rat ASIC3 (rASIC3) produced only minor are activated by extracellular protons and are differences in channel function, and replacing involved in a wide range of physiological and nmrASIC3 sequence with that of rASIC3 did not pathophysiological processes, including pain and produce a proton-sensitive ion channel.