Clinical Factors That Influence the Cellular Responses of Saphenous
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analyses of Allele-Specific Gene Expression in Highly Divergent
ARTICLES Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance James J Crowley1,10, Vasyl Zhabotynsky1,10, Wei Sun1,2,10, Shunping Huang3, Isa Kemal Pakatci3, Yunjung Kim1, Jeremy R Wang3, Andrew P Morgan1,4,5, John D Calaway1,4,5, David L Aylor1,9, Zaining Yun1, Timothy A Bell1,4,5, Ryan J Buus1,4,5, Mark E Calaway1,4,5, John P Didion1,4,5, Terry J Gooch1,4,5, Stephanie D Hansen1,4,5, Nashiya N Robinson1,4,5, Ginger D Shaw1,4,5, Jason S Spence1, Corey R Quackenbush1, Cordelia J Barrick1, Randal J Nonneman1, Kyungsu Kim2, James Xenakis2, Yuying Xie1, William Valdar1,4, Alan B Lenarcic1, Wei Wang3,9, Catherine E Welsh3, Chen-Ping Fu3, Zhaojun Zhang3, James Holt3, Zhishan Guo3, David W Threadgill6, Lisa M Tarantino7, Darla R Miller1,4,5, Fei Zou2,11, Leonard McMillan3,11, Patrick F Sullivan1,5,7,8,11 & Fernando Pardo-Manuel de Villena1,4,5,11 Complex human traits are influenced by variation in regulatory DNA through mechanisms that are not fully understood. Because regulatory elements are conserved between humans and mice, a thorough annotation of cis regulatory variants in mice could aid in further characterizing these mechanisms. Here we provide a detailed portrait of mouse gene expression across multiple tissues in a three-way diallel. Greater than 80% of mouse genes have cis regulatory variation. Effects from these variants influence complex traits and usually extend to the human ortholog. Further, we estimate that at least one in every thousand SNPs creates a cis regulatory effect. -

Expression and Function of FERMT Genes in Colon Carcinoma Cells
ANTICANCER RESEARCH 33: 167-174 (2013) Expression and Function of FERMT Genes in Colon Carcinoma Cells KENJI KIRIYAMA1,2,3, YOSHIHIKO HIROHASHI1, TOSHIHIKO TORIGOE1, TERUFUMI KUBO1, YASUAKI TAMURA1, TAKAYUKI KANASEKI1, AKARI TAKAHASHI1, EMIRI NAKAZAWA1, ERI SAKA1, CHARLOTTE RAGNARSSON1, MUNEHIDE NAKATSUGAWA1, SATOKO INODA1,2, HIROKO ASANUMA4, HIDEO TAKASU5, TADASHI HASEGAWA4, TAKAHIRO YASOSHIMA3, KOICHI HIRATA2 and NORIYUKI SATO1 Department of 1Pathology, 2Surgery Ist and 4Surgical Pathology, Sapporo Medical University School of Medicine, Sapporo, Japan; 3Department of Surgery, Shinsapporo Keiaikai Hospital, Sapporo, Japan; 5Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan Abstract. Invasion into the matrix is one of hallmarks of Colon carcinoma is a major malignancy, with a high malignant diseases and is the first step for tumor metastasis. mortality rate. In the process of tumorigenesis, tumor cells Thus, analysis of the molecular mechanisms of invasion is undergo multiple steps of genetic events (1), and multiple essential to overcome tumor cell invasion. In the present study, steps are also required for the cells to obtain several different we screened for colon carcinoma-specific genes using a cDNA phenotypes. Tissue invasion and metastasis are hallmarks microarray database of colon carcinoma tissues and normal that distinguish malignant from benign diseases (2). Several colon tissues, and we found that fermitin family member-1 classes of proteins are involved in the process of tissue (FERMT1) is overexpressed in colon carcinoma cells. FRRMT1, invasion; however, the exact molecular mechanisms of FERMT2 and FERMT3 expression was investigated in colon invasion remain unclear. carcinoma cells. Reverse transcription polymerase chain Fermitin family member (FERMT) genes include FERMT1, reaction (RT-PCR) analysis revealed that only FERMT1 had FERMT2 and FERMT3, and these genes have been reported cancer cell-specific expression. -

Bayesian GWAS and Network Analysis Revealed New Candidate Genes for Number of Teats in Pigs
J Appl Genetics DOI 10.1007/s13353-014-0240-y ANIMAL GENETICS • ORIGINAL PAPER Bayesian GWAS and network analysis revealed new candidate genes for number of teats in pigs L. L. Verardo & F. F. Silva & L. Varona & M. D. V. Resende & J. W. M. Bastiaansen & P. S. Lopes & S. E. F. Guimarães Received: 7 March 2014 /Revised: 27 May 2014 /Accepted: 23 July 2014 # Institute of Plant Genetics, Polish Academy of Sciences, Poznan 2014 Abstract The genetic improvement of reproductive traits comparisons based on deviance posterior distribution indicat- such as the number of teats is essential to the success of the ed the superiority of Gaussian model. In general, our results pig industry. As opposite to most SNP association studies that suggest the presence of 19 significant SNPs, which mapped consider continuous phenotypes under Gaussian assumptions, 13 genes. Besides, we predicted gene interactions through this trait is characterized as a discrete variable, which could networks that are consistent with the mammals known breast potentially follow other distributions, such as the Poisson. biology (e.g., development of prolactin receptor signaling, and Therefore, in order to access the complexity of a counting cell proliferation), captured known regulation binding sites, random regression considering all SNPs simultaneously as and provided candidate genes for that trait (e.g., TINAGL1 covariate under a GWAS modeling, the Bayesian inference and ICK). tools become necessary. Currently, another point that deserves to be highlighted in GWAS is the genetic dissection of com- Keywords Counting data . Genes . Reproductive traits . SNP plex phenotypes through candidate genes network derived association from significant SNPs. -
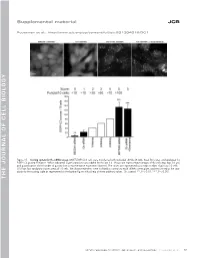
Thejournalofcellb Io Logy
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

Identification of Potential Key Genes and Pathway Linked with Sporadic Creutzfeldt-Jakob Disease Based on Integrated Bioinformatics Analyses
medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification of potential key genes and pathway linked with sporadic Creutzfeldt-Jakob disease based on integrated bioinformatics analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract Sporadic Creutzfeldt-Jakob disease (sCJD) is neurodegenerative disease also called prion disease linked with poor prognosis. The aim of the current study was to illuminate the underlying molecular mechanisms of sCJD. The mRNA microarray dataset GSE124571 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

Linkage and Association Studies in African
中国科技论文在线 http://www.paper.edu.cn Molecular Psychiatry (2006),1–12 & 2006 Nature Publishing Group All rights reserved 1359-4184/06 $30.00 www.nature.com/mp ORIGINAL ARTICLE Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence MD Li1, D Sun1,2, X-Y Lou1, J Beuten1, TJ Payne3 and JZ Ma4 1Department of Psychiatry and Neurobehavioral Sciences, University of Virginia, Charlottesville, VA, USA; 2Department of Animal Genetics and Breeding and Key Laboratory of Animal Genetics and Breeding of the Ministry of Agriculture, China Agricultural University, Beijing, PR China; 3ACT Center for Tobacco Treatment, Education and Research, University of Mississippi Medical Center, Jackson, MS, USA and 4Department of Public Health Sciences, University of Virginia, Charlottesville, VA, USA Our previous linkage study demonstrated that the 9q22–q23 chromosome region showed a ‘suggestive’ linkage to nicotine dependence (ND) in the Framingham Heart Study population. In this study, we provide further evidence for the linkage of this region to ND in an independent sample. Within this region, the gene encoding Src homology 2 domain-containing transform- ing protein C3 (SHC3) represents a plausible candidate for association with ND, assessed by smoking quantity (SQ), the Heaviness of Smoking Index (HSI) and the Fagerstro¨ m Test for ND (FTND). We utilized 11 single-nucleotide polymorphisms within SHC3 to examine the association with ND in 602 nuclear families of either African-American (AA) or European- American (EA) origin. Individual SNP-based analysis indicated three SNPs for AAs and one for EAs were significantly associated with at least one ND measure. -

Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
International Journal of Molecular Sciences Review Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling Maria Carmela Annunziata 1, Melania Parisi 1, Gabriella Esposito 2 , Gabriella Fabbrocini 1, Rosario Ammendola 2 and Fabio Cattaneo 2,* 1 Department of Clinical Medicine and Surgery, School of Medicine, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (M.C.A.); [email protected] (M.P.); [email protected] (G.F.) 2 Department of Molecular Medicine and Medical Biotechnology, School of Medicine,, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (G.E.); [email protected] (R.A.) * Correspondence: [email protected]; Fax: +39-081-7464-359 Received: 5 May 2020; Accepted: 25 May 2020; Published: 27 May 2020 Abstract: FPR1, FPR2, and FPR3 are members of Formyl Peptides Receptors (FPRs) family belonging to the GPCR superfamily. FPR2 is a low affinity receptor for formyl peptides and it is considered the most promiscuous member of this family. Intracellular signaling cascades triggered by FPRs include the activation of different protein kinases and phosphatase, as well as tyrosine kinase receptors transactivation. Protein kinases and phosphatases act coordinately and any impairment of their activation or regulation represents one of the most common causes of several human diseases. Several phospho-sites has been identified in protein kinases and phosphatases, whose role may be to expand the repertoire of molecular mechanisms of regulation or may be necessary for fine-tuning of switch properties. We previously performed a phospho-proteomic analysis in FPR2-stimulated cells that revealed, among other things, not yet identified phospho-sites on six protein kinases and one protein phosphatase. -

Genetic Network Identifies Novel Pathways Contributing To
Albright et al. BMC Medical Genomics 2014, 7:51 http://www.biomedcentral.com/1755-8794/7/51 RESEARCH ARTICLE Open Access Genetic network identifies novel pathways contributing to atherosclerosis susceptibility in the innominate artery Jody Albright3, Pamela M Quizon3, Aldons J Lusis4,5,6 and Brian J Bennett1,2,3* Abstract Background: Atherosclerosis, the underlying cause of cardiovascular disease, results from both genetic and environmental factors. Methods: In the current study we take a systems-based approach using weighted gene co-expression analysis to identify a candidate pathway of genes related to atherosclerosis. Bioinformatic analyses are performed to identify candidate genes and interactions and several novel genes are characterized using in-vitro studies. Results: We identify 1 coexpression module associated with innominate artery atherosclerosis that is also enriched for inflammatory and macrophage gene signatures. Using a series of bioinformatics analysis, we further prioritize the genes in this pathway and identify Cd44 as a critical mediator of the atherosclerosis. We validate our predictions generated by the network analysis using Cd44 knockout mice. Conclusion: These results indicate that alterations in Cd44 expression mediate inflammation through a complex transcriptional network involving a number of previously uncharacterized genes. Keywords: Atherosclerosis, Co-expression network, inflammation Background act in part by affecting local inflammation in the vessel Cardiovascular disease (CVD) is the leading cause of wall [3-5]. death in the United States and the incidence of CVD is Inflammation is a fundamental biological process that rapidly increasing in other countries [1]. A common cause provides protection from infection, but when dysregulated, of cardiovascular disease is atherosclerosis, a pathological acts as a source of chronic disease. -

STRIPAK Complexes in Cell Signaling and Cancer
Oncogene (2016), 1–9 © 2016 Macmillan Publishers Limited All rights reserved 0950-9232/16 www.nature.com/onc REVIEW STRIPAK complexes in cell signaling and cancer Z Shi1,2, S Jiao1 and Z Zhou1,3 Striatin-interacting phosphatase and kinase (STRIPAK) complexes are striatin-centered multicomponent supramolecular structures containing both kinases and phosphatases. STRIPAK complexes are evolutionarily conserved and have critical roles in protein (de) phosphorylation. Recent studies indicate that STRIPAK complexes are emerging mediators and regulators of multiple vital signaling pathways including Hippo, MAPK (mitogen-activated protein kinase), nuclear receptor and cytoskeleton remodeling. Different types of STRIPAK complexes are extensively involved in a variety of fundamental biological processes ranging from cell growth, differentiation, proliferation and apoptosis to metabolism, immune regulation and tumorigenesis. Growing evidence correlates dysregulation of STRIPAK complexes with human diseases including cancer. In this review, we summarize the current understanding of the assembly and functions of STRIPAK complexes, with a special focus on cell signaling and cancer. Oncogene advance online publication, 15 February 2016; doi:10.1038/onc.2016.9 INTRODUCTION in the central nervous system and STRN4 is mostly abundant in Recent proteomic studies identified a group of novel multi- the brain and lung, whereas STRN3 is ubiquitously expressed in 5–9 component complexes named striatin (STRN)-interacting phos- almost all tissues. STRNs share a -

Mining Genes in Type 2 Diabetic Islets and Finding Gold
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Cell Metabolism Previews Fischer, C., Mazzone, M., Jonckx, B., and Carme- Same´ n, E., Lu, L., et al. (2012). Nature 490, Takamoto, I., Sasako, T., et al. (2011). Cell Metab. liet, P. (2008). Nat. Rev. Cancer 8, 942–956. 426–430. 13, 294–307. Hagberg, C.E., Falkevall, A., Wang, X., Larsson, E., Karpanen, T., Bry, M., Ollila, H.M., Seppa¨ nen- Poesen, K., Lambrechts, D., Van Damme, P., Huusko, J., Nilsson, I., van Meeteren, L.A., Samen, Laakso, T., Liimatta, E., Leskinen, H., Kivela¨ , R., Dhondt, J., Bender, F., Frank, N., Bogaert, E., E., Lu, L., Vanwildemeersch, M., et al. (2010). Helkamaa, T., Merentie, M., Jeltsch, M., et al. Claes, B., Heylen, L., Verheyen, A., et al. (2008). Nature 464, 917–921. (2008). Circ. Res. 103, 1018–1026. J. Neurosci. 28, 10451–10459. Hagberg, C.E., Mehlem, A., Falkevall, A., Muhl, L., Kubota, T., Kubota, N., Kumagai, H., Yamaguchi, Samuel, V.T., and Shulman, G.I. (2012). Cell 148, Fam, B.C., Ortsa¨ ter, H., Scotney, P., Nyqvist, D., S., Kozono, H., Takahashi, T., Inoue, M., Itoh, S., 852–871. Mining Genes in Type 2 Diabetic Islets and Finding Gold Decio L. Eizirik1,* and Miriam Cnop1,2 1Laboratory of Experimental Medicine, Medical Faculty 2Division of Endocrinology, Erasmus Hospital Universite Libre de Bruxelles (ULB), 1000 Brussels, Belgium *Correspondence: [email protected] http://dx.doi.org/10.1016/j.cmet.2012.10.012 Pancreatic b cell failure is central in the pathogenesis of type 2 diabetes (T2D), but the mechanisms involved remain unclear. -

Kindlin-3 Mutation in Mesenchymal Stem Cells Results in Enhanced Chondrogenesis
bioRxiv preprint doi: https://doi.org/10.1101/578690; this version posted March 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Kindlin-3 Mutation in Mesenchymal Stem Cells Results in Enhanced Chondrogenesis Bethany A. Kerr1,2,3, Lihong Shi2, Alexander H. Jinnah3, Jeffrey S. Willey3,4, Donald P. Lennon5, Arnold I. Caplan5, Tatiana V. Byzova1 1 Department of Molecular Cardiology, Joseph J. Jacobs Center for Thrombosis and Vascular Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, 44195 2 Department of Cancer Biology and Comprehensive Cancer Center, Wake Forest School of Medicine, Winston-Salem, NC 27157 3 Department of Orthopaedic Surgery, Wake Forest School of Medicine, Winston-Salem, NC 27157 4 Department of Radiation Biology, Wake Forest School of Medicine, Winston-Salem, NC 27157 5 Skeletal Research Center, Department of Biology, Case Western Reserve University, Cleveland, Ohio, 44106 Running Title: Kindlin-3 regulates chondrogenesis Address correspondence to: Bethany Kerr, Ph.D., Wake Forest School of Medicine, 1 Medical Center Blvd, Winston-Salem, NC, 27157. Telephone: 336-716-0320; Twitter: @BethanyKerrLab; e-mail: [email protected]; ORCID: 0000-0002-2995-7549 Number of Figures: 7 Number of Tables: 1 bioRxiv preprint doi: https://doi.org/10.1101/578690; this version posted March 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity.