Rediscovery of Nycticebus Coucang Insularis Robinson, 1917
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Data Confirm the Presence of Nycticebus Bengalensis on Langkawi Island, Malaysia
BIODIVERSITAS ISSN: 1412-033X Volume 20, Number 4, April 2019 E-ISSN: 2085-4722 Pages: 1115-1120 DOI: 10.13057/biodiv/d200424 Molecular data confirm the presence of Nycticebus bengalensis on Langkawi Island, Malaysia BADRUL MUNIR MD-ZAIN1,, KHAIRUL SYA’ADAH MOHHOYUA1, NOR RAHMAN AIFAT1, EHWAN NGADI1, NORSHAQINAH AYOB1, JEFFRINE JAPNING ROVIE-RYAN2, AHMAD AMPENG3, ABD RAHMAN MOHD-RIDWAN1,4, MARY E BLAIR5, MUHAMMAD ABU BAKAR ABDUL-LATIFF6 1School of Environmental and Natural Resource Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia. 43600, Bangi, Selangor, Malaysia. Tel.: +60-389-213200, email: [email protected], [email protected] 2Department of Wildlife and National Park (DWNP) Peninsular Malaysia, Km 10 Jalan Cheras, 56100 Kuala Lumpur, Malaysia 3Sarawak Forest Department, Wisma Sumber Alam Jalan Stadium, 93660 Petra Jaya, Kuching, Sarawak, Malaysia 4Centre for Pre-University Studies, Universiti Malaysia Sarawak,94300 Kota Samarahan, Sarawak, Malaysia 5Center for Biodiversity and Conservation, American Museum of Natural History, Central Park West & 79th St, New York, NY 10024, United States 6Centre of Research for Sustainable Uses of Natural Resources (CoR-SUNR), Faculty of Applied Sciences and Technology, Universiti Tun Hussein Onn Malaysia (Pagoh Campus). 84000, Muar, Johor, Malaysia Manuscript received: 23 February 2019. Revision accepted: 24 March 2019. Abstract. Md-Zain BM, Mohhoyua KS, Aifat NR, Ngadi E, Ayob N, Rovie-Ryan JJ, Ampeng A, Mohd-Ridwan AR, Blair ME, Abdul-Latiff MAB. 2019. Molecular data confirm the presence of Nycticebus bengalensis on Langkawi Island, Malaysia. Biodiversitas 20: 1115- 1120. Recent taxonomic reviews have stated the possibility of Bengal Slow Loris (Nycticebus bengalensis) presence in the Northern part of the Malay Peninsula. -

Pongo Abelii)
UvA-DARE (Digital Academic Repository) Orangutan diet: lessons from and for the wild Hardus, M.E. Publication date 2012 Link to publication Citation for published version (APA): Hardus, M. E. (2012). Orangutan diet: lessons from and for the wild. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:29 Sep 2021 4 Behavioral, Ecological, and Evolutionary Aspects of Meat-Eating by Sumatran Orangutans (Pongo abelii) Madeleine E. Hardus, Adriano R. Lameira, Astri Zulfa, S. Suci Utami Atmoko, Han de Vries, Serge A. Wich Abstract Meat-eating is an important aspect of human evolution, but how meat became a substantial component of the human diet is still poorly understood. Meat eating in our closest relatives, the great apes, may provide insight into the emergence of this trait, but most existing data are for chimpanzees. -

Downloaded from Brill.Com09/27/2021 09:14:05PM Via Free Access 218 Rode-Margono & Nekaris – Impact of Climate and Moonlight on Javan Slow Lorises
Contributions to Zoology, 83 (4) 217-225 (2014) Impact of climate and moonlight on a venomous mammal, the Javan slow loris (Nycticebus javanicus Geoffroy, 1812) Eva Johanna Rode-Margono1, K. Anne-Isola Nekaris1, 2 1 Oxford Brookes University, Gipsy Lane, Headington, Oxford OX3 0BP, UK 2 E-mail: [email protected] Keywords: activity, environmental factors, humidity, lunarphobia, moon, predation, temperature Abstract Introduction Predation pressure, food availability, and activity may be af- To secure maintenance, survival and reproduction, fected by level of moonlight and climatic conditions. While many animals adapt their behaviour to various factors, such nocturnal mammals reduce activity at high lunar illumination to avoid predators (lunarphobia), most visually-oriented nocturnal as climate, availability of resources, competition, preda- primates and birds increase activity in bright nights (lunarphilia) tion, luminosity, habitat fragmentation, and anthropo- to improve foraging efficiency. Similarly, weather conditions may genic disturbance (Kappeler and Erkert, 2003; Beier influence activity level and foraging ability. We examined the 2006; Donati and Borgognini-Tarli, 2006). According response of Javan slow lorises (Nycticebus javanicus Geoffroy, to optimal foraging theory, animal behaviour can be seen 1812) to moonlight and temperature. We radio-tracked 12 animals as a trade-off between the risk of being preyed upon in West Java, Indonesia, over 1.5 years, resulting in over 600 hours direct observations. We collected behavioural and environmen- and the fitness gained from foraging (Charnov, 1976). tal data including lunar illumination, number of human observ- Perceived predation risk assessed through indirect cues ers, and climatic factors, and 185 camera trap nights on potential that correlate with the probability of encountering a predators. -

The Illegal Exploitation of the Javan Leopard (
Nature Conservation 43: 25–39 (2021) A peer-reviewed open-access journal doi: 10.3897/natureconservation.43.59399 RESEARCH ARticlE https://natureconservation.pensoft.net Launched to accelerate biodiversity conservation The illegal exploitation of the Javan Leopard (Panthera pardus melas) and Sunda Clouded Leopard (Neofelis diardi) in Indonesia Lalita Gomez1,2, Chris R. Shepherd1 1 Monitor Conservation Research Society, Big Lake, Canada 2 Oxford Wildlife Trade Research Group, Oxford Brookes University, Oxford, UK Corresponding author: Chris R. Shepherd ([email protected]) Academic editor: M. Auliya | Received 6 October 2020 | Accepted 15 January 2021 | Published 22 March 2021 http://zoobank.org/17D9AAB6-8A94-4B5A-932F-6633FAD5D42B Citation: Gomez L, Shepherd CR (2021) The illegal exploitation of the Javan Leopard (Panthera pardus melas) and Sunda Clouded Leopard (Neofelis diardi) in Indonesia. Nature Conservation 43: 25–39. https://doi.org/10.3897/ natureconservation.43.59399 Abstract Indonesia is home to the Javan Leopard (Panthera pardus melas) and the Sunda Clouded Leopard (Neofelis diardi), both of which are threatened by habitat loss, human-wildlife conflict issues and the illegal wildlife trade. Leopards and clouded leopards are threatened by the illegal wildlife trade across their range, how- ever, very little is known of the illegal trade in these two species in Indonesia, or of the efforts made to tackle this crime. Both the Javan Leopard and Sunda Clouded Leopard are protected species in Indonesia and both species are listed in Appendix I of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), meaning commercial international trade is generally prohibited. To better understand the trade, and efforts to end this trade, we collected records of seizures and prosecutions relating to Javan Leopards and Sunda Clouded Leopards in Indonesia for the period 2011–2019. -

Slow Loris Forest Protector Teacher's Pack
2 www.nocturama.org https://www.facebook.com/pages/Little-Fireface-Project/ littlefi[email protected] INTRODUCTION Welcome! Welcome to the Slow Loris—Forest Protector’s Teacher’s Pack and Learning Exercise Book. The purpose of this short booklet is to explain how to use the book and to help to reinforce its message through a series of fun and easy-to- We at the Little Fireface Project (LFP) our so glad you are using this teacher’s use exercises. pack. First we want to tell you about ourselves and our passion about one of the world’s most unique little primates. LFP is a UK Charity based out of Ox- ford Brookes University set up to help save the slow loris through studying In this pack, you will find the following materials to help you and your students them in the wild and through education projects. explore the story of two night-active (nocturnal) primates, slow lorises: a moth- er (Tereh—speedy) and her young son (Bunga—flower). Tereh lovingly teaches Why the slow loris and why a charity dedicated to one group of animals? Well, her son the life skills he needs to be a grown-up slow loris. At the same time, the eight species of slow lorises, found only in Asia, are facing a tough time. Bunga learns that by doing his job in the forest, he helps the forest to grow, They are threatened for many reasons beyond the habitat loss causing many of while helping protect the crops grown by people. Asia’s species to go extinct. -

World's Most Endangered Primates
Primates in Peril The World’s 25 Most Endangered Primates 2016–2018 Edited by Christoph Schwitzer, Russell A. Mittermeier, Anthony B. Rylands, Federica Chiozza, Elizabeth A. Williamson, Elizabeth J. Macfie, Janette Wallis and Alison Cotton Illustrations by Stephen D. Nash IUCN SSC Primate Specialist Group (PSG) International Primatological Society (IPS) Conservation International (CI) Bristol Zoological Society (BZS) Published by: IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), Bristol Zoological Society (BZS) Copyright: ©2017 Conservation International All rights reserved. No part of this report may be reproduced in any form or by any means without permission in writing from the publisher. Inquiries to the publisher should be directed to the following address: Russell A. Mittermeier, Chair, IUCN SSC Primate Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, VA 22202, USA. Citation (report): Schwitzer, C., Mittermeier, R.A., Rylands, A.B., Chiozza, F., Williamson, E.A., Macfie, E.J., Wallis, J. and Cotton, A. (eds.). 2017. Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. 99 pp. Citation (species): Salmona, J., Patel, E.R., Chikhi, L. and Banks, M.A. 2017. Propithecus perrieri (Lavauden, 1931). In: C. Schwitzer, R.A. Mittermeier, A.B. Rylands, F. Chiozza, E.A. Williamson, E.J. Macfie, J. Wallis and A. Cotton (eds.), Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018, pp. 40-43. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. -

SILVERY GIBBON PROJECT Newsletterthe Page 1 September 2013 SILVERY GIBBON PROJECT
SILVERY GIBBON PROJECT NEWSLETTERThe Page 1 September 2013 SILVERY GIBBON PROJECT PO BOX 335 COMO 6952 WESTERN AUSTRALIA Website: www.silvery.org.au E-mail: [email protected] Phone: 0438992325 September 2013 Agile Gibbons, Siamangs, Sunbears, Clouded Leopards and even Sumatran Tigers, the area lies PRESIDENT’S REPORT adjacent to a larger protected forest and is also the location of Kalaweit Conservation Centre. We Dear Members and Friends hope to be able to provide additional support to this important project into the future. Keep an eye We are excited to report that the release of on our Facebook page for updates on camera trap Sadewa and Kiki in June went very well and they images from Supayang. continue to thrive in the forest. They are proving a little challenging for the monitoring team to keep Back in Perth, the SGP team is gearing up for our up with but thanks to their morning call the team Art Auction which will be held on October 26. are still able to locate them most days. Please Once again we have secured some amazing read the update on their release on page 4. pieces and the generosity of artists is truly inspiring. Be sure to get your tickets early as this event may sell out. We also have some exciting projects underway with Wildlife Asia. You can find out more about our crowd funding project on page 5. It was certainly an honour to be present at the release, which was the culmination of many years hard work for the Java Gibbon Centre (JGC) team. -

Hibernation in Pygmy Lorises (Nycticebus Pygmaeus) – What Does It Mean?
Vietnamese Journal of Primatology (2017) vol.2(5), 51-57 Hibernation in pygmy lorises (Nycticebus pygmaeus) – what does it mean? Ulrike Streicher1,3, Julia Nowack2, Gabrielle Stalder2, Christian Walzer2, Tilo Nadler3 and Thomas Ruf2 1 Current address: Cascades Raptor Center, Eugene, USA 2 University of Veterinary Medicine, Research Institute of Wildlife Ecology, Department of Integrative Biology and Evolution, Vienna, Austria, Savoyenstr. 1, 110 Vienna, Austria 3 Endangered Primate Rescue Center, Cuc Phương National Park, Nho Quan District, Ninh Bình Province, Vietnam Corresponding author: Ulrike Streicher <[email protected]> Key words: South-East Asia, primate, torpor, multiday torpor, pygmy loris, hibernation Summary Torpor use in primates appeared to be restricted to African species and was only recently discovered in a species from Asia, the pygmy loris (Nycticebus pygmaeus). This finding has considerable implications for our perception of torpor in this mammal group and demonstrates that torpor is probably more widespread in mammals than commonly thought. This article summarizes the current knowledge on the use of torpor in the pygmy loris and places it into the context of ongoing research on this topic. Hiện tượng ngủ đông ở loài culi nhỏ (Nycticebus pygmaeus) – Ý nghĩa là gì? Tóm tắt Hiện tượng ngủ đông ở các loài linh trưởng được cho rằng chỉ tồn tại ở một số loài linh trưởng ở Châu Phi. Gần đây hiện tượng này được khám phá ở một loài linh trưởng ở Châu Á, loài culi nhỏ (Nycticebus pygmaeus). Phát hiện mới này có thể thay đổi nhận thức của chúng ta về hiện tượng ngủ đông ở nhóm thú này và nó cũng minh chứng rằng hiện tượng ngủ đông có thể phổ biến ở nhiều loài thú khác hơn những gì chúng ta thường nghĩ. -

New Species of Slow Loris Discovered in Borneo
New Species Of Slow Loris Discovered In Borneo - December 17, 2012 Asian Scientist Magazine | Science, Technology and Medicine News Updates From Asia - http://www.asianscientist.com New Species Of Slow Loris Discovered In Borneo December 17, 2012 http://www.asianscientist.com/2012/12/in-the-lab/new-species-of-slow-loris-nycticebus-kayan-in- borneo-2012/ AsianScientist (Dec. 17, 2012) - Scientists have discovered three new species of the elusive slow loris in Borneo, rare among primates for having a toxic bite. The team's analysis of the primate's distinctive facial fur markings, published in the American Journal of Primatology, reveals the existence of one entirely new species, while two of species, previously considered as possible sub-species, are being officially recognized as unique. "Technological advances have improved our knowledge about the diversity of several nocturnal mammals," said Rachel Munds from the University of Missouri Columbia. "Historically many species went unrecognized as they were falsely lumped together as one species. While the number of recognized primate species has doubled in the past 25 years some nocturnal species remain hidden to science." The slow loris (Nycticebus) is a primate genus closely related to the lemur. Found across Southeast Asia, from Bangladesh and China's Yunnan province to the island of Borneo, the slow loris is rare among primates for having a toxic bite, and is rated as Vulnerable or Endangered on the IUCN Red List. Slow lorises are recognized by their unique fur coloration on the body and face, yet while traits such as fur patterns are often used to distinguish between species; nocturnal species are cryptic in coloration and have less obvious external differences. -

Fine Scale Habitat and Movement Patterns of Javan Slow Loris ( Nycticebus Javanicus )In Cipaganti, West Java, Indonesia
Fine scale habitat and movement patterns of javan slow loris ( Nycticebus javanicus )in Cipaganti, West Java, Indonesia Lina Fransson Degree project in biology, Master of science (2 years), 2018 Examensarbete i biologi 45 hp till masterexamen, 2018 Biology Education Centre, Uppsala University, and Department of Ecology and Genetics/ Animal Ecology, in cooperation with the Little Fireface Project Supervisors: Frank Johansson and Marie Sigaud External opponent: Josefine Stångberg Abstract....................................................................................................................................... 1 Introduction ................................................................................................................................ 2 Study questions ....................................................................................................................... 4 Materials and methods................................................................................................................ 5 Study area ............................................................................................................................... 5 “The species studied” in relation to its movement ................................................................. 6 Field work ............................................................................................................................... 8 Vegetation and habitat study ............................................................................................... -

Clouded Leopard Common Names: Kung (Bhotia), Amchita (Nepali), Pungmar (Lepcha)
Neofelis nebulosa Family: Clouded leopard Common names: Kung (Bhotia), Amchita (Nepali), Pungmar (Lepcha) Classification: Kingdom: Animalia Phylum: Chordata Class: Mammalia Order: Carniv Family: Felidae Genus: Neofelis Species: nebulosa Profile: Despite its name, the clouded leopard is not closely related to the leopard. The smallest of the big cats, the species is sufficiently distinct from other members of the Felidae family to be placed in a separate genus – Neofelis, which is originating from two words, neo meaning "new", and felis meaning "small cat". The common name is derived from its characteristic coat pattern. The ochre coat is patterned with grey elliptical clouds edged with black, which enable it to camouflage itself in the forest. In size, the species is intermediate between the large and small cats, with a body length of 60-110 cm and weighing between 11-20 kg. The head and the legs are spotted. The animal has two broad bars on its neck, and stripes on its cheeks. It has a very long tail marked with broken black rings and short legs with large broad paws, which give it a rather heavy appearance. The paws are adapted for an arboreal lifestyle. Among the felids, it has the longest canines in proportion to its skull size. The clouded leopard occupies the highest position in the tropical rainforest food chain. Lifespan: Average 11 years (in the wild). They live longer in captivity. Distribution: The clouded leopard is found along the Himalayan foothills, from Nepal, through mainland Southeast Asia and into China. Although historically the species enjoyed a wide distribution in China, current distribution in the region is largely unknown. -
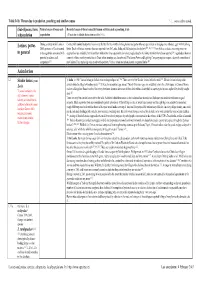
Table 14 B: Threat Due to Predation, Poaching and Similar Causes 1, 2,
Table 14 b: Threat due to predation, poaching and similar causes 1, 2, ... : source, author quoted. (Sub-)Species, form, Natural causes of losses such Recorded causes of threat caused by human activities such as poaching, trade subpopulation as predation (Threat due to habitat destruction see table 14 c) Lorises, pottos During a survey in Sri Lanka in Contact with uninsulated power lines is may be fatal for the slowly climbing lorises and pottos who are specialized on bridging over substrate gaps with their long 2001, presence of Loris seemed limbs. Death of lorises on power lines are reported from Sri Lanka, India and Malaysia (see also below) 207, 66, 211. From Africa, no data concerning pottos or in general to be negatively associated with angwantibos are available, but the problem with power lines apparently also exists, regular deaths of colobus monkeys have been reported 208, regarded as the main potential predators and cause of colobus monkey mortalities in Diani; other monkeys are also affected. The Kenya Power and Lighting Company engineers spent a day with coworkers of competitors 211. the Colobus Trust exploring ways to solve the problem. Victims of electrocution showed amputated limbs 209. Asian lorises L I Slender lorises, genus In India, in 1981 "lorises belong to India's most endangered species" 116. There are very few Slender lorises left in the wild 121. Slender lorises belong to the Loris animals found in illegal ownership (pets) 110. It was also a popular cage animal. Twenty-five years ago, one could buy a loris for a few rupees in Chennai Moore market or Bangalore Russel market.