Cross%Lineage,Hybridization,In,The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Palm Haven Garden Plant List - 2010
PALM HAVEN GARDEN PLANT LIST - 2010 BOTANICAL NAME COMMON NAME Achillea millefolium 'Paprika' Paprika yarrow Arctostaphylos glauca big berry manzanita Arctostaphylos pajaroensis Pajaro manzanita Arctostaphylos uva-ursi bearberry Arctostaphylos uva-ursi 'San Bruno Mountain' San Bruno Mountain bearberry Aristolochia californica California pipevine Artemisia pycnocephala coastal sagewort Baccharis pilularis 'Twin Peaks' Twin Peaks dwarf coyote brush Berberis aquifolium 'Golden Abundance' Golden Abundance Oregon-grape Carex spissa San Diego sedge Ceanothus 'Joyce Coulter' Joyce Coulte wild lilac Ceanothus arboreus feltleaf ceanothus Ceanothus gloriosus var exaltatus 'Emily Brown' Emily Brown glory brush Dendromecon harfordii Channel Island tree poppy Epilobium canum California fuchsia Erigeron glaucus seaside daisy Eriogonum fasciculatum California buckwheat Eriogonum latifolium coast buckwheat Eriogonum umbellatum sulfur flower Eriophyllum confertiflorum golden yarrow Heteromeles arbutifolia toyon Heterotheca sessilifolora false goldenaster Iris douglasiana Douglas iris Leymus condensatus 'Canyon Prince' Canyon Prince giant wild rye Mimulus aurantiacus sticky monkeyflower Monardella villosa coyote mint Mulhenbergia rigens deer grass Penstemon heterophyllus foothill penstemon Physocarpus capitatus ninebark Rhamnus californica 'Ed Holm' Ed Holm coffeeberry Rhamnus californica 'Mound San Bruno' Mound San Bruno coffeeberry Salvia clevelandii Cleveland sage Salvia mellifera 'Shirley's Creeper' Shirley's Creepe sage Salvia sonomensis 'Bee's Bliss' Bee's Bliss sage Satureja douglasii yerba buena Trichostema lanatum wolly blue curls Typha angustifolia narrow-leaved cattail Verbena lilacina lilac verbena Vitis californica 'Roger's Red' Roger's Red California wild grape. -
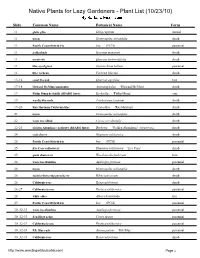
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Qty Size Name 6 1G Abies Bracteata 10 1G Abutilon Palmeri 1 1G Acaena Pinnatifida Var
REGIONAL PARKS BOTANIC GARDEN, TILDEN REGIONAL PARK, BERKELEY, CALIFORNIA Celebrating 76 years of growing California native plants: 1940-2016 **FINAL**PLANT SALE LIST **FINAL** (9/30/2016 @ 6:00 PM) visit: www.nativeplants.org for the most up to date plant list FALL PLANT SALE OF CALIFORNIA NATIVE PLANTS SATURDAY, OCTOBER 1, 2016 PUBLIC SALE: 10:00 AM TO 3:00 PM MEMBERS ONLY SALE: 9:00 AM TO 10:00 AM MEMBERSHIPS ARE AVAILABLE AT THE ENTRY TO THE SALE AT 8:30 AM Qty Size Name 6 1G Abies bracteata 10 1G Abutilon palmeri 1 1G Acaena pinnatifida var. californica 18 1G Achillea millefolium 10 4" Achillea millefolium - Black Butte 28 4" Achillea millefolium 'Island Pink' 8 4" Achillea millefolium 'Rosy Red' - donated by Annie's Annuals 2 4" Achillea millefolium 'Sonoma Coast' 7 4" Acmispon (Lotus) argophyllus var. argenteus 9 1G Actea rubra f. neglecta (white fruits) 25 4" Adiantum x tracyi (A. jordanii x A. aleuticum) 5 1G Aesculus californica 1 2G Agave shawii var. shawii 2 1G Agoseris grandiflora 8 1G Alnus incana var. tenuifolia 2 2G Alnus incana var. tenuifolia 5 4" Ambrosia pumila 5 1G Amelanchier alnifolia var. semiintegrifolia 9 1G Anemopsis californica 5 1G Angelica hendersonii 3 1G Angelica tomentosa 1 1G Apocynum androsaemifolium x Apocynum cannabinum 7 1G Apocynum cannabinum 5 1G Aquilegia formosa 2 4" Aquilegia formosa 4 4" Arbutus menziesii 2 1G Arctostaphylos andersonii 2 1G Arctostaphylos auriculata 3 1G Arctostaphylos 'Austin Griffith' 11 1G Arctostaphylos bakeri 5 1G Arctostaphylos bakeri 'Louis Edmunds' 2 1G Arctostaphylos canescens 2 1G Arctostaphylos canescens subsp. -

Other Botanical Resource Assessment
USDA Forest Service Tahoe National Forest District Yuba River Ranger District OTHER BOTANICAL RESOURCE ASSESSMENT Yuba Project 08/01/2017 Prepared by: Date: Courtney Rowe, District Botanist TABLE OF CONTENTS 1 TNF Watch List Botanical Species ........................................................................................................ 1 1.1 Introduction ................................................................................................................................ 1 1.2 Summary of Analysis Procedure .................................................................................................. 2 1.3 Project Compliance ..................................................................................................................... 2 2 Special Status Plant Communities ....................................................................................................... 5 2.1 Introduction ................................................................................................................................ 5 2.2 Project Compliance ..................................................................................................................... 5 3 Special Management Designations ..................................................................................................... 6 3.1 Introduction ................................................................................................................................ 6 3.2 Project Compliance .................................................................................................................... -

Propagation and Cultivation of Arctostaphylos in Relation to the Environment in Its Natural Habitat 291
Propagation and Cultivation of Arctostaphylos in Relation to the Environment in its Natural Habitat 291 Propagation and Cultivation of Arctostaphylos in Relation to the Environment in its Natural Habitat in California, U.S.A.© Lucy Hart' School of Horticulture, Royal Botanic Gardens Kew, Richmond, Surrey TW9 3AB U.K. INTRODUCTION The Mary Helliar Travel Scholarship helped to fund a visit to California to study native plants in their natural habitats and in cultivation. Throughout my study I observed Arctostaphylos, commonly known as manzanita, growing naturally and was able to relate the natural habitats to cultivation conditions in botanic gardens and commercial nurseries where I learnt about the propagation and production of members of the genus. Arctostaphylos is a fundamental genus to California, found almost exclusively in the state, with different species occupying a range of habitats. It is a member of the Ericaceae and is closely related to Arbutus, sharing the same subfamily, Arbutoideae. The generic name is derived from two Greek words — arktos meaning bear and stuphule, a grape. The common name, manzanita (popularly used in California today) is Spanish for "little apple" from the appearance of its berry. There are approximately 60 species, of which several have many subspecies due to frequent hybridisations within the genus (Stuart and Sawyer, 2001). This can make identification difficult in areas where species ranges overlap. Schmidt (1973), a manzanita enthusiast, describes her excitement regarding the future possibilities for more horticultural forms from the natural hybridisations, as a "tantalising prospect." KEY HORTICULTURAL FEATURES The genus includes many forms of evergreen, woody shrubs ranging from low, prostrate, mat-forming types to a few which approach tree size. -

Qty Size Name Price 10 1G Abies Bracteata 12.00 $ 15 1G Abutilon
REGIONAL PARKS BOTANIC GARDEN, TILDEN REGIONAL PARK, BERKELEY, CALIFORNIA Celebrating 78 years of growing California native plants: 1940-2018 **PRELIMINARY**PLANT SALE LIST **PRELIMINARY** Preliminary Plant Sale List 9/29/2018 visit: www.nativeplants.org for the most up to date plant list, updates are posted until 10/5 FALL PLANT SALE OF CALIFORNIA NATIVE PLANTS SATURDAY, October 6, 2018 PUBLIC SALE: 10:00 AM TO 3:00 PM MEMBERS ONLY SALE: 9:00 AM TO 10:00 AM MEMBERSHIPS ARE AVAILABLE AT THE ENTRY TO THE SALE AT 8:30 AM Qty Size Name Price 10 1G Abies bracteata $ 12.00 15 1G Abutilon palmeri $ 11.00 1 1G Acer circinatum $ 10.00 3 5G Acer circinatum $ 40.00 8 1G Acer macrophyllum $ 9.00 10 1G Achillea millefolium 'Calistoga' $ 8.00 25 4" Achillea millefolium 'Island Pink' OUR INTRODUCTION! $ 5.00 28 1G Achillea millefolium 'Island Pink' OUR INTRODUCTION! $ 8.00 6 1G Actea rubra f. neglecta (white fruits) $ 9.00 3 1G Adenostoma fasciculatum $ 10.00 1 4" Adiantum aleuticum $ 10.00 6 1G Adiantum aleuticum $ 13.00 10 4" Adiantum shastense $ 10.00 4 1G Adiantum x tracyi $ 13.00 2 2G Aesculus californica $ 12.00 1 4" Agave shawii var. shawii $ 8.00 1 1G Agave shawii var. shawii $ 15.00 4 1G Allium eurotophilum $ 10.00 3 1G Alnus incana var. tenuifolia $ 8.00 4 1G Amelanchier alnifolia var. semiintegrifolia $ 9.00 8 2" Anemone drummondii var. drummondii $ 4.00 9 1G Anemopsis californica $ 9.00 8 1G Apocynum cannabinum $ 8.00 2 1G Aquilegia eximia $ 8.00 15 4" Aquilegia formosa $ 6.00 11 1G Aquilegia formosa $ 8.00 10 1G Aquilegia formosa 'Nana' $ 8.00 Arabis - see Boechera 5 1G Arctostaphylos auriculata $ 11.00 2 1G Arctostaphylos auriculata - large inflorescences from Black Diamond $ 11.00 1 1G Arctostaphylos bakeri $ 11.00 15 1G Arctostaphylos bakeri 'Louis Edmunds' $ 11.00 2 1G Arctostaphylos canescens subsp. -

International Ecological Classification Standard
INTERNATIONAL ECOLOGICAL CLASSIFICATION STANDARD: TERRESTRIAL ECOLOGICAL CLASSIFICATIONS Groups and Macrogroups of Washington June 26, 2015 by NatureServe (modified by Washington Natural Heritage Program on January 16, 2016) 600 North Fairfax Drive, 7th Floor Arlington, VA 22203 2108 55th Street, Suite 220 Boulder, CO 80301 This subset of the International Ecological Classification Standard covers vegetation groups and macrogroups attributed to Washington. This classification has been developed in consultation with many individuals and agencies and incorporates information from a variety of publications and other classifications. Comments and suggestions regarding the contents of this subset should be directed to Mary J. Russo, Central Ecology Data Manager, NC <[email protected]> and Marion Reid, Senior Regional Ecologist, Boulder, CO <[email protected]>. Copyright © 2015 NatureServe, 4600 North Fairfax Drive, 7th floor Arlington, VA 22203, U.S.A. All Rights Reserved. Citations: The following citation should be used in any published materials which reference ecological system and/or International Vegetation Classification (IVC hierarchy) and association data: NatureServe. 2015. International Ecological Classification Standard: Terrestrial Ecological Classifications. NatureServe Central Databases. Arlington, VA. U.S.A. Data current as of 26 June 2015. Restrictions on Use: Permission to use, copy and distribute these data is hereby granted under the following conditions: 1. The above copyright notice must appear in all documents and reports; 2. Any use must be for informational purposes only and in no instance for commercial purposes; 3. Some data may be altered in format for analytical purposes, however the data should still be referenced using the citation above. Any rights not expressly granted herein are reserved by NatureServe. -

Arctostaphylos Hispidula, Gasquet Manzanita
Conservation Assessment for Gasquet Manzanita (Arctostaphylos hispidula) Within the State of Oregon Photo by Clint Emerson March 2010 U.S.D.A. Forest Service Region 6 and U.S.D.I. Bureau of Land Management Interagency Special Status and Sensitive Species Program Author CLINT EMERSON is a botanist, USDA Forest Service, Rogue River-Siskiyou National Forest, Gold Beach and Powers Ranger District, Gold Beach, OR 97465 TABLE OF CONTENTS Disclaimer 3 Executive Summary 3 List of Tables and Figures 5 I. Introduction 6 A. Goal 6 B. Scope 6 C. Management Status 7 II. Classification and Description 8 A. Nomenclature and Taxonomy 8 B. Species Description 9 C. Regional Differences 9 D. Similar Species 10 III. Biology and Ecology 14 A. Life History and Reproductive Biology 14 B. Range, Distribution, and Abundance 16 C. Population Trends and Demography 19 D. Habitat 21 E. Ecological Considerations 25 IV. Conservation 26 A. Conservation Threats 26 B. Conservation Status 28 C. Known Management Approaches 32 D. Management Considerations 33 V. Research, Inventory, and Monitoring Opportunities 35 Definitions of Terms Used (Glossary) 39 Acknowledgements 41 References 42 Appendix A. Table of Known Sites in Oregon 45 2 Disclaimer This Conservation Assessment was prepared to compile existing published and unpublished information for the rare vascular plant Gasquet manzanita (Arctostaphylos hispidula) as well as include observational field data gathered during the 2008 field season. This Assessment does not represent a management decision by the U.S. Forest Service (Region 6) or Oregon/Washington BLM. Although the best scientific information available was used and subject experts were consulted in preparation of this document, it is expected that new information will arise. -

Site Assessemnt (PDF)
Site Assessment Report Scotts Valley Hotel SCOTTS VALLEY, SANTA CRUZ COUNTY, CALIFORNIA December 29, 2014 Prepared by: On behalf of: Johnson Marigot Consulting, LLC City Ventures, LLC Cameron Johnson Mr. Jason Bernstein 88 North Hill Drive, Suite C 444 Spear Street, Suite 200 Brisbane, California 94005 San Francisco, California 94105 1 Table Of Contents SECTION 1: Environmental Setting ................................................................................... 4 A. Project Location ........................................................................................................................... 4 B. Surrounding Land Use ................................................................................................................ 4 C. Study Area Topography and Hydrology ............................................................................... 4 D. Study Area Soil .............................................................................................................................. 5 E. Vegetation Types .......................................................................................................................... 5 SECTION 2: Methods ............................................................................................................... 7 A. Site Visit .......................................................................................................................................... 7 B. Study Limits .................................................................................................................................. -

Oak Resources Management Plan
EXHIBIT A El Dorado County Oak Resources Managenient Plan September 2017 El Dorado County Community Development Agency Long Range Planning Division 2850 Fairlane Court, Placerville, CA 95667 OAK RESOURCES MANAGEMENT PLAN Table of Contents 1.0 Introduction ................................................................................................................... l 1.1 Purpose ...................................................................................................................1 1.2 Goals and Objectives of Plan .................................................................................2 1.3 Oak Resources in El Dorado County .....................................................................3 1.3.1 Oak Woodlands ........................................................................................... .3 1.3 .2 Oak Trees .................................................................................................... .4 1.4 Economic Activity, Land, and Ecosystem Values of Oak Resources ....................4 1.5 State-level Regulations ...........................................................................................4 2.0 Oak Resources Impact Mitigation Requirements .........................................................6 2.1 Applicability, Exemptions and Mitigation Reductions ..........................................6 2.1.1 Single-Family Lot Exemption...................................................................... 6 2.1.2 Fire Safe Activities Exemption ....................................................................6 -

Verdura® Native Planting
Abronia maritime Abronia maritima is a species of sand verbena known by the common name red (Coastal) sand verbena. This is a beach-adapted perennial plant native to the coastlines of southern California, including the Channel Islands, and northern Baja California. Abronia villosa Abronia villosa is a species of sand-verbena known by the common name desert (Inland) sand-verbena. It is native to the deserts of the southwestern United States and northern Mexico and the southern California and Baja coast. Adenostoma Adenostoma fasciculatum (chamise or greasewood) is a flowering plant native to fasciculatum California and northern Baja California. This shrub is one of the most widespread (Coastal/Inland) plants of the chaparral biome. Adenostoma fasciculatum is an evergreen shrub growing to 4m tall, with dry-looking stick-like branches. The leaves are small, 4– 10 mm long and 1mm broad with a pointed apex, and sprout in clusters from the branches. Arctostaphylos Arctostaphylos uva-ursi is a plant species of the genus Arctostaphylos (manzanita). uva-ursi Its common names include kinnikinnick and pinemat manzanita, and it is one of (Coastal/Inland) several related species referred to as bearberry. Arctostaphylos Arctostaphylos edmundsii, with the common name Little Sur manzanita, is a edmundsii species of manzanita. This shrub is endemic to California where it grows on the (Coastal/Inland) coastal bluffs of Monterey County. Arctostaphylos Arctostaphylos hookeri is a species of manzanita known by the common name hookeri Hooker's manzanita. Arctostaphylos hookeri is a low shrub which is variable in (Coastal/Inland) appearance and has several subspecies. The Arctostaphylos hookeri shrub is endemic to California where its native range extends from the coastal San Francisco Bay Area to the Central Coast. -

Cavitation Resistance Among 26 Chaparral Species of Southern California
Ecological Monographs, 77(1), 2007, pp. 99–115 Ó 2007 by the Ecological Society of America CAVITATION RESISTANCE AMONG 26 CHAPARRAL SPECIES OF SOUTHERN CALIFORNIA 1,4 2 1 3 ANNA L. JACOBSEN, R. BRANDON PRATT, FRANK W. EWERS, AND STEPHEN D. DAVIS 1Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824-1312 USA 2Department of Biology, California State University, Bakersfield, 14 SCI, 9001 Stockdale Highway, Bakersfield, California 93311 USA 3Natural Science Division, Pepperdine University, 24255 Pacific Coast Highway, Malibu, California 90263 USA Abstract. Resistance to xylem cavitation depends on the size of xylem pit membrane pores and the strength of vessels to resist collapse or, in the case of freezing-induced cavitation, conduit diameter. Altering these traits may impact plant biomechanics or water transport efficiency. The evergreen sclerophyllous shrub species, collectively referred to as chaparral, which dominate much of the mediterranean-type climate region of southern California, have been shown to display high cavitation resistance (pressure potential at 50% loss of hydraulic conductivity; P50). We examined xylem functional and structural traits associated with more negative P50 in stems of 26 chaparral species. We correlated raw-trait values, without phylogenetic consideration, to examine current relationships between P50 and these xylem traits. Additionally, correlations were examined using phylogenetic independent contrasts (PICs) to determine whether evolutionary changes in these xylem traits correlate with changes in P50. Co-occurring chaparral species widely differ in their P50 (À0.9 to À11.0 MPa). Species experiencing the most negative seasonal pressure potential (Pmin) had the highest resistance to xylem cavitation (lowest P50). Decreased P50 was associated with increased xylem density, stem mechanical strength (modulus of rupture), and transverse fiber wall area when both raw values and PICs were analyzed.