Mbd4 Inactivation Increases C3T Transition Mutations and Promotes Gastrointestinal Tumor Formation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Developmental Delay and the Methyl Binding Genes H Turner, F Macdonald, S Warburton, F Latif, T Webb
1of3 ELECTRONIC LETTER J Med Genet: first published as 10.1136/jmg.40.2.e13 on 1 February 2003. Downloaded from Developmental delay and the methyl binding genes H Turner, F MacDonald, S Warburton, F Latif, T Webb ............................................................................................................................. J Med Genet 2003;40:e13(http://www.jmedgenet.com/cgi/content/full/40/2/e13) he report by Amir et al1 that Rett syndrome (RS) is associ- children referred with a clinical diagnosis of Angelman ated with mutations in the MECP2 gene permitted labora- syndrome (AS) but without an abnormality in 15q11q13 tory diagnosis of this devastating yet common neurode- showed that 4/46 of the girls in one case8 and 5/40 in the T 9 velopmental disorder. Hitherto the paucity of familial cases of other actually had Rett syndrome. Of these nine probands, the syndrome and the failure to identify the syndrome in only one was later found to have a clinical presentation incon- males despite fairly wide clinical criteria had defined it as an sistent with the laboratory diagnosis. This may not be surpris- X linked dominant disorder with male lethality.2 Soon, ing given that in very small girls the two syndromes may however, reports from the few families in which RS is present with overlapping clinical features and so be difficult to segregating showed that male family members who inherited differentiate on clinical grounds alone. the same mutation in the MECP2 gene as their affected female Familial cases of developmental handicap -

Epigenetic Regulation of DNA Repair Genes and Implications for Tumor Therapy ⁎ ⁎ Markus Christmann , Bernd Kaina
Mutation Research-Reviews in Mutation Research xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Mutation Research-Reviews in Mutation Research journal homepage: www.elsevier.com/locate/mutrev Review Epigenetic regulation of DNA repair genes and implications for tumor therapy ⁎ ⁎ Markus Christmann , Bernd Kaina Department of Toxicology, University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany ARTICLE INFO ABSTRACT Keywords: DNA repair represents the first barrier against genotoxic stress causing metabolic changes, inflammation and DNA repair cancer. Besides its role in preventing cancer, DNA repair needs also to be considered during cancer treatment Genotoxic stress with radiation and DNA damaging drugs as it impacts therapy outcome. The DNA repair capacity is mainly Epigenetic silencing governed by the expression level of repair genes. Alterations in the expression of repair genes can occur due to tumor formation mutations in their coding or promoter region, changes in the expression of transcription factors activating or Cancer therapy repressing these genes, and/or epigenetic factors changing histone modifications and CpG promoter methylation MGMT Promoter methylation or demethylation levels. In this review we provide an overview on the epigenetic regulation of DNA repair genes. GADD45 We summarize the mechanisms underlying CpG methylation and demethylation, with de novo methyl- TET transferases and DNA repair involved in gain and loss of CpG methylation, respectively. We discuss the role of p53 components of the DNA damage response, p53, PARP-1 and GADD45a on the regulation of the DNA (cytosine-5)- methyltransferase DNMT1, the key enzyme responsible for gene silencing. We stress the relevance of epigenetic silencing of DNA repair genes for tumor formation and tumor therapy. -

Deficiency in DNA Mismatch Repair of Methylation Damage Is a Major
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.18.388108; this version posted November 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Deficiency in DNA mismatch repair of methylation damage is a major 2 mutational process in cancer 3 1 1 1 1 1,* 4 Hu Fang , Xiaoqiang Zhu , Jieun Oh , Jayne A. Barbour , Jason W. H. Wong 5 6 1School of Biomedical Sciences, Li Ka Shing Faculty of Medicine, The University of 7 Hong Kong, Hong Kong Special Administrative Region 8 9 *Correspondence: [email protected] 10 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.11.18.388108; this version posted November 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Abstract 2 DNA mismatch repair (MMR) is essential for maintaining genome integrity with its 3 deficiency predisposing to cancer1. MMR is well known for its role in the post- 4 replicative repair of mismatched base pairs that escape proofreading by DNA 5 polymerases following cell division2. Yet, cancer genome sequencing has revealed that 6 MMR deficient cancers not only have high mutation burden but also harbour multiple 7 mutational signatures3, suggesting that MMR has pleotropic effects on DNA repair. The 8 mechanisms underlying these mutational signatures have remained unclear despite 9 studies using a range of in vitro4,5 and in vivo6 models of MMR deficiency. -

Involvement of MBD4 Inactivation in Mismatch Repair-Deficient Tumorigenesis
Involvement of MBD4 inactivation in mismatch repair-deficient tumorigenesis The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Tricarico, R., S. Cortellino, A. Riccio, S. Jagmohan-Changur, H. van der Klift, J. Wijnen, D. Turner, et al. 2015. “Involvement of MBD4 inactivation in mismatch repair-deficient tumorigenesis.” Oncotarget 6 (40): 42892-42904. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26318553 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 40 Involvement of MBD4 inactivation in mismatch repair-deficient tumorigenesis Rossella Tricarico1, Salvatore Cortellino2, Antonio Riccio3, Shantie Jagmohan- Changur4, Heleen Van der Klift5, Juul Wijnen5, David Turner6, Andrea Ventura7, Valentina Rovella8, Antonio Percesepe9, Emanuela Lucci-Cordisco10, Paolo Radice11, Lucio Bertario11, Monica Pedroni12, Maurizio Ponz de Leon12, Pietro Mancuso1,13, Karthik Devarajan14, Kathy Q. Cai15, Andres J.P. Klein-Szanto15, Giovanni Neri10, Pål Møller16, Alessandra Viel17, Maurizio Genuardi10, Riccardo Fodde4, Alfonso Bellacosa1 1Cancer Epigenetics and Cancer Biology Programs, Fox Chase Cancer Center, Philadelphia, Pennsylvania, United States 2IFOM-FIRC Institute of Molecular Oncology, Milan, -

Mismatch Repair
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair Gerald T. Marsischky, Nicole Filosi, Michael F. Kane, and Richard Kolodner I Charles A. Dana Division of Human Cancer Genetics, Dana-Farber Cancer Institute, Boston, Massachusetts 09.115 USA; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115 USA Saccharomyces cerevisiae encodes six genes, MSH1-6, which encode proteins related to the bacterial MutS protein. In this study the role of MSH2, MSH3, and MSH6 in mismatch repair has been examined by measuring the rate of accumulating mutations and mutation spectrum in strains containing different combinations of rash2, rash3, and rash6 mutations and by studying the physical interaction between the MSH2 protein and the MSH3 and MSH6 proteins. The results indicate that S. cerevisiae has two pathways of MSH2-dependent mismatch repair: one that recognizes single-base mispairs and requires MSH2 and MSH6, and a second that recognizes insertion/deletion mispairs and requires a combination of either MSH2 and MSH6 or MSH2 and MSH3. The redundancy of MSH3 and MSH6 explains the greater prevalence of brash2 mutations in HNPCC families and suggests how the role of brash3 and hmsh6 mutations in cancer susceptibility could be analyzed. [Key Words: Cancer; mutagenesis; mismatch repair; routS; MSH2; MSH3; MSH6; Saccharomyces cerevisiae] Received October 17, 1995; revised version accepted January 17, 1996. DNA mismatch repair plays a number of roles in the cell al. 1993; Umar et al. 1994; Boyer et al. -

RESEARCH ARTICLE Regulatory Network of Micrornas, Target
DOI:http://dx.doi.org/10.7314/APJCP.2015.16.2.475 Regulatory Network of MicroRNAs, Target Genes, Transcription Factors and Host Genes in Endometrial Cancer RESEARCH ARTICLE Regulatory Network of MicroRNAs, Target Genes, Transcription Factors and Host Genes in Endometrial Cancer Lu-Chen Xue1,3, Zhi-Wen Xu2,3*, Kun-Hao Wang2,3, Ning Wang2,3, Xiao-Xu Zhang1, Shang Wang2,3 Abstract Genes and microRNAs (miRNAs) have important roles in human oncology. However, most of the biological factors are reported in disperse form which makes it hard to discover the pathology. In this study, genes and miRNAs involved in human endometrial cancer(EC) were collected and formed into regulatory networks following their interactive relations, including miRNAs targeting genes, transcription factors (TFs) regulating miRNAs and miRNAs included in their host genes. Networks are constructed hierarchically at three levels: differentially expressed, related and global. Among the three, the differentially expressed network is the most important and fundamental network that contains the key genes and miRNAs in EC. The target genes, TFs and miRNAs are differentially expressed in EC so that any mutation in them may impact on EC development. Some key pathways in networks were highlighted to analyze how they interactively influence other factors and carcinogenesis. Upstream and downstream pathways of the differentially expressed genes and miRNAs were compared and analyzed. The purpose of this study was to partially reveal the deep regulatory mechanisms in EC using a new method that combines comprehensive genes and miRNAs together with their relationships. It may contribute to cancer prevention and gene therapy of EC. -

DNA Excision Repair Proteins and Gadd45 As Molecular Players for Active DNA Demethylation
Cell Cycle ISSN: 1538-4101 (Print) 1551-4005 (Online) Journal homepage: http://www.tandfonline.com/loi/kccy20 DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation Dengke K. Ma, Junjie U. Guo, Guo-li Ming & Hongjun Song To cite this article: Dengke K. Ma, Junjie U. Guo, Guo-li Ming & Hongjun Song (2009) DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation, Cell Cycle, 8:10, 1526-1531, DOI: 10.4161/cc.8.10.8500 To link to this article: http://dx.doi.org/10.4161/cc.8.10.8500 Published online: 15 May 2009. Submit your article to this journal Article views: 135 View related articles Citing articles: 92 View citing articles Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=kccy20 Download by: [University of Pennsylvania] Date: 27 April 2017, At: 12:48 [Cell Cycle 8:10, 1526-1531; 15 May 2009]; ©2009 Landes Bioscience Perspective DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation Dengke K. Ma,1,2,* Junjie U. Guo,1,3 Guo-li Ming1-3 and Hongjun Song1-3 1Institute for Cell Engineering; 2Department of Neurology; and 3The Solomon Snyder Department of Neuroscience; Johns Hopkins University School of Medicine; Baltimore, MD USA Abbreviations: DNMT, DNA methyltransferases; PGCs, primordial germ cells; MBD, methyl-CpG binding protein; NER, nucleotide excision repair; BER, base excision repair; AP, apurinic/apyrimidinic; SAM, S-adenosyl methionine Key words: DNA demethylation, Gadd45, Gadd45a, Gadd45b, Gadd45g, 5-methylcytosine, deaminase, glycosylase, base excision repair, nucleotide excision repair DNA cytosine methylation represents an intrinsic modifica- silencing of gene activity or parasitic genetic elements (Fig. -

The Properties of Msh2–Msh6 ATP Binding Mutants Suggest a Signal
ARTICLE cro Author’s Choice The properties of Msh2–Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair Received for publication, August 19, 2018, and in revised form, September 17, 2018 Published, Papers in Press, September 20, 2018, DOI 10.1074/jbc.RA118.005439 William J. Graham, V‡, Christopher D. Putnam‡§, and X Richard D. Kolodner‡¶ʈ**1 From the ‡Ludwig Institute for Cancer Research San Diego, Departments of §Medicine and ¶Cellular and Molecular Medicine, ʈMoores-UCSD Cancer Center, and **Institute of Genomic Medicine, University of California School of Medicine, San Diego, La Jolla, California 92093-0669 Edited by Patrick Sung DNA mismatch repair (MMR) corrects mispaired DNA bases synthesis (1–6). MMR also acts on some forms of chemically and small insertion/deletion loops generated by DNA replica- damaged DNA bases as well as on mispairs present in hetero- tion errors. After binding a mispair, the eukaryotic mispair rec- duplex DNA intermediates formed during recombination ognition complex Msh2–Msh6 binds ATP in both of its nucle- (1–6). MMR proteins also act in a pathway that suppresses otide-binding sites, which induces a conformational change recombination between homologous but divergent DNAs resulting in the formation of an Msh2–Msh6 sliding clamp that (1–6) and function in a DNA damage response pathway for releases from the mispair and slides freely along the DNA. How- different types of DNA-damaging agents, including some che- ever, the roles that Msh2–Msh6 sliding clamps play in MMR motherapeutic agents (2). As a result of the role that MMR plays remain poorly understood. -
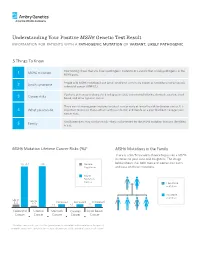
MSH6 Genetic Test Result Information for Patients with a Pathogenic Mutation Or Variant, Likely Pathogenic
Understanding Your Positive MSH6 Genetic Test Result information for patients with a pathogenic mutation or variant, likely pathogenic 5 Things To Know Your testing shows that you have a pathogenic mutation or a variant that is likely pathogenic in the 1 MSH6 mutation MSH6 gene. People with MSH6 mutations have Lynch syndrome, previously known as hereditary non-polyposis 2 Lynch syndrome colorectal cancer (HNPCC). You have an increased chance to develop colorectal, endometrial/uterine, stomach, ovarian, small 3 Cancer risks bowel, and other types of cancer. There are risk management options to detect cancer early or lower the risk to develop cancer. It is 4 What you can do important to discuss these options with your doctor, and decide on a plan that best manages your cancer risks. Family members may also be at risk – they can be tested for the MSH6 mutation that was identified 5 Family in you. MSH6 Mutation Lifetime Cancer Risks (%)* MSH6 Mutations in the Family There is a 50/50 random chance to pass on a MSH6 mutation to your sons and daughters. The image 20-44 ~44 General below shows that both men and women can carry Population and pass on these mutations. MSH6 Mutation Carrier Has MSH6 mutation No MSH6 mutation Up to Up to 5.5 Increased Increased Increased 2.7 <1 <1 <1 Colorectal Uterine Stomach Ovarian Small Bowel Cancer Cancer Cancer Cancer Cancer *The above cancer risks represent the typical range for individuals with a mutation in this gene. If available, cancer risks specific to the mutation found in you will be provided in your results report. -

Lynch Syndrome (MLH1, MSH2, MSH6, PMS2, EPCAM) - Update 2012
European Journal of Human Genetics (2013) 21; doi:10.1038/ejhg.2012.164 & 2013 Macmillan Publishers Limited All rights reserved 1018-4813/13 www.nature.com/ejhg CLINICAL UTILITY GENE CARD UPDATE Clinical utility gene card for: Lynch syndrome (MLH1, MSH2, MSH6, PMS2, EPCAM) - update 2012 Nils Rahner*,1, Verena Steinke2, Brigitte Schlegelberger3, Francois Eisinger4, Pierre Hutter5 and Sylviane Olschwang4 European Journal of Human Genetics (2013) 21, doi:10.1038/ejhg.2012.164; published online 15 August 2012 Update to: European Journal of Human Genetics (2010) 18, 1069; doi:10.1038/ejhg.2009.232; published online 27 January 2010 1. DISEASE CHARACTERISTICS MLH1;lossofMSH2/MSH6–analysisofMSH2; isolated loss of 1.1 Name of the disease (synonyms) MSH6 – analysis of MSH6; isolated loss of PMS2 – analysis of Lynch syndrome/HNPCC. PMS2). Germline analysis should include search for point mutations and large genomic deletions/duplications/insertions 1.2 OMIM# of the disease (e.g., by pre-screening (DHPLC), direct sequencing on gDNA or 276300, 613244. cDNA level, MLPA including promoter regions and EPCAM gene, Southern blot analysis). Mutation analysis of PMS2 should include 1.3 Name of the analysed genes or DNA/chromosome segments the analysis on RNA.9 Germline MLH1 promoter methylation MLH1, MSH2, MSH6, PMS2, and EPCAM. characterisation by MSP, bisulphite pyrosequencing, or MLPA (useful for diagnostic purpose, not for predictive testing). 1.4 OMIM# of the gene(s) MLH1 (120436), MSH2 (609309), MSH6 (600678), PMS2 (600259), EPCAM (185535). 1.7 Analytical validation Confirmation of mutation in an independent biological sample of the 1.5 Mutational spectrum index case or an affected relative. -

Constitutional Mismatch Repair Deficiency Syndrome
Constitutional mismatch repair deficiency syndrome Description Constitutional mismatch repair deficiency (CMMRD) syndrome is a rare disorder that greatly increases the risk of developing one or more types of cancer in children and young adults. The cancers that most commonly occur in CMMRD syndrome are cancers of the colon (large intestine) and rectum (collectively referred to as colorectal cancer), brain, and blood (leukemia or lymphoma). Almost all people with CMMRD syndrome develop cancer before age 18, generally in late childhood. The age of diagnosis varies depending on the cancer type; brain cancers, leukemia, and lymphomas tend to occur at younger ages than colorectal cancer in people with CMMRD syndrome. It is estimated that 20 to 40 percent of people with CMMRD syndrome who develop cancer will develop another cancer later in life. People with CMMRD syndrome may develop multiple noncancerous (benign) growths ( adenomas) in the colon that are likely to become cancerous (malignant) over time. Brain cancers in CMMRD syndrome often involve certain cells called glial cells, causing gliomas or glioblastomas. The most common blood cancer in CMMRD syndrome is called non-Hodgkin lymphoma, which affects white blood cells. Other cancers that can occur in CMMRD syndrome include cancers of the small intestine, urinary tract, or uterine lining (endometrial cancer). Many people with CMMRD syndrome develop features similar to those that occur in a condition called neurofibromatosis type 1. These features include changes in skin coloring (pigmentation), which are characterized by one or more flat patches on the skin that are darker than the surrounding area (café-au-lait spots). Some affected individuals have freckling or patches of skin that are unusually light in color (hypopigmented). -

MSH6 and PMS2 Mutation Positive Australian Lynch Syndrome Families
Talseth-Palmer et al. Hereditary Cancer in Clinical Practice 2010, 8:5 http://www.hccpjournal.com/content/8/1/5 RESEARCH Open Access MSH6 and PMS2 mutation positive Australian Lynch syndrome families: novel mutations, cancer risk and age of diagnosis of colorectal cancer Bente A Talseth-Palmer1,2*, Mary McPhillips3, Claire Groombridge4, Allan Spigelman5, Rodney J Scott1,2,3 Abstract Background: Approximately 10% of Lynch syndrome families have a mutation in MSH6 and fewer families have a mutation in PMS2. It is assumed that the cancer incidence is the same in families with mutations in MSH6 as in families with mutations in MLH1/MSH2 but that the disease tends to occur later in life, little is known about families with PMS2 mutations. This study reports on our findings on mutation type, cancer risk and age of diagnosis in MSH6 and PMS2 families. Methods: A total of 78 participants (from 29 families) with a mutation in MSH6 and 7 participants (from 6 families) with a mutation in PMS2 were included in the current study. A database of de-identified patient information was analysed to extract all relevant information such as mutation type, cancer incidence, age of diagnosis and cancer type in this Lynch syndrome cohort. Cumulative lifetime risk was calculated utilising Kaplan-Meier survival analysis. Results: MSH6 and PMS2 mutations represent 10.3% and 1.9%, respectively, of the pathogenic mutations in our Australian Lynch syndrome families. We identified 26 different MSH6 and 4 different PMS2 mutations in the 35 families studied. We report 15 novel MSH6 and 1 novel PMS2 mutations.