Next-Generation Sequencing Analysis of Receptor-Type Tyrosine Kinase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
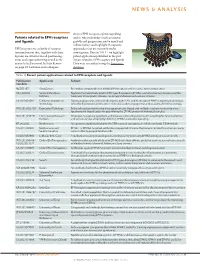
Patents Related to EPH Receptors and Ligands
NEWS & ANALYSIS discuss EPH receptor–ephrin signalling Patents related to EPH receptors and its role in disorders such as tumour and ligands growth and progression, nerve injury and inflammation, and highlight therapeutic EPH receptors are a family of receptor approaches that are currently under tyrosine kinases that, together with their investigation. Here in TABLE 1 we highlight ligands, are involved in cell positioning, patent applications published in the past tissue and organ patterning as well as the 3 years related to EPH receptors and ligands. control of cell survival. In their Review Data were researched using the Espacenet on page 39, Lackman and colleagues database. Table 1 | Recent patent applications related to EPH receptors and ligands Nature Reviews | Drug Discovery Publication Applicants Subject numbers NZ 581397 AstraZeneca Pyrimidine compounds that inhibit EPH receptors and are useful for treating cancer HK 1108702 Sanford-Burnham Peptides that selectively bind to EPH type-B receptors (EPHBs); useful for tumour imaging and the Institute treatment of neoplastic disease, neurological disease and vascular disease US 2013091591 California Institute of During angiogenesis, arterial cells express ephrin B2, and its receptor EPHB4 is expressed on venous Technology cells; this distinction can be used in methods to alter angiogenesis and to assess the effect of drugs WO 2013052710 Expression Pathology Selected reaction monitoring mass spectrometry-based and multiple reaction monitoring mass spectrometry-based assays for quantifying -

Human FLT4 / VEGFR3 ELISA Kit (ARG82047)
Product datasheet [email protected] ARG82047 Package: 96 wells Human FLT4 / VEGFR3 ELISA Kit Store at: 4°C Summary Product Description Human FLT4 / VEGFR3 ELISA Kit is an Enzyme Immunoassay kit for the quantification of Human FLT4 / VEGFR3 in serum, plasma and cell culture supernatants. Tested Reactivity Hu Tested Application ELISA Target Name FLT4 / VEGFR3 Conjugation HRP Conjugation Note Substrate: TMB and read at 450 nm. Sensitivity 78 pg/ml Sample Type Serum, plasma and cell culture supernatants. Standard Range 156 - 10000 pg/ml Sample Volume 100 µl Alternate Names FLT-4; FLT41; Vascular endothelial growth factor receptor 3; VEGFR3; VEGFR-3; PCL; Tyrosine-protein kinase receptor FLT4; LMPH1A; EC 2.7.10.1; Fms-like tyrosine kinase 4 Application Instructions Assay Time 4.5 hours Properties Form 96 well Storage instruction Store the kit at 2-8°C. Keep microplate wells sealed in a dry bag with desiccants. Do not expose test reagents to heat, sun or strong light during storage and usage. Please refer to the product user manual for detail temperatures of the components. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol FLT4 Gene Full Name fms-related tyrosine kinase 4 Background This gene encodes a tyrosine kinase receptor for vascular endothelial growth factors C and D. The protein is thought to be involved in lymphangiogenesis and maintenance of the lymphatic endothelium. Mutations in this gene cause hereditary lymphedema type IA. [provided by RefSeq, Jul 2008] Function Tyrosine-protein kinase that acts as a cell-surface receptor for VEGFC and VEGFD, and plays an essential role in adult lymphangiogenesis and in the development of the vascular network and the cardiovascular system during embryonic development. -

A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma
414 Original Article A Review of VEGF/VEGFR-Targeted Therapeutics for Recurrent Glioblastoma David A. Reardon, MDa,b; Scott Turner, MDc; Katherine B. Peters, MD, PhDc; Annick Desjardins, MDc; Sridharan Gururangan, MDa,b; John H. Sampson, MD, PhDa; Roger E. McLendon, MDd; James E. Herndon II, PhDe; Lee W. Jones, PhDf; John P. Kirkpatrick, MD, PhDf; Allan H. Friedman, MDa; James J. Vredenburgh, MDc; Darell D. Bigner, MD, PhDd; and Henry S. Friedman, MDa,b; Durham, North Carolina Key Words ability effect of VEGF inhibitors, the Radiologic Assessment in Neu- Glioblastoma, angiogenesis, vascular endothelial growth factor, ro-Oncology (RANO) criteria were recently implemented to bet- malignant glioma ter assess response among patients with glioblastoma. Although bevacizumab improves survival and quality of life, eventual tumor progression is the norm. Better understanding of resistance mech- Abstract anisms to VEGF inhibitors and identification of effective therapy Glioblastoma, the most common primary malignant brain tumor after bevacizumab progression are currently a critical need for pa- among adults, is a highly angiogenic and deadly tumor. Angiogen- tients with glioblastoma. (JNCCN 2011;9:414–427) esis in glioblastoma, driven by hypoxia-dependent and indepen- dent mechanisms, is primarily mediated by vascular endothelial growth factor (VEGF), and generates blood vessels with distinctive features. The outcome for patients with recurrent glioblastoma is poor because of ineffective therapies. However, recent encourag- ing rates of radiographic response and progression-free survival, Malignant gliomas, including the most common sub- and adequate safety, led the FDA to grant accelerated approval of type of glioblastoma, are rapidly growing destructive tu- bevacizumab, a humanized monoclonal antibody against VEGF, for mors that extensively invade locally but rarely metasta- the treatment of recurrent glioblastoma in May 2009. -

Original Article ERBB3, IGF1R, and TGFBR2 Expression Correlate With
Am J Cancer Res 2018;8(5):792-809 www.ajcr.us /ISSN:2156-6976/ajcr0077452 Original Article ERBB3, IGF1R, and TGFBR2 expression correlate with PDGFR expression in glioblastoma and participate in PDGFR inhibitor resistance of glioblastoma cells Kang Song1,2*, Ye Yuan1,2*, Yong Lin1,2, Yan-Xia Wang1,2, Jie Zhou1,2, Qu-Jing Gai1,2, Lin Zhang1,2, Min Mao1,2, Xiao-Xue Yao1,2, Yan Qin1,2, Hui-Min Lu1,2, Xiang Zhang1,2, You-Hong Cui1,2, Xiu-Wu Bian1,2, Xia Zhang1,2, Yan Wang1,2 1Department of Pathology, Institute of Pathology and Southwest Cancer Center, Southwest Hospital, Third Military Medical University, Chongqing 400038, China; 2Key Laboratory of Tumor Immunology and Pathology of Ministry of Education, Chongqing 400038, China. *Equal contributors. Received April 6, 2018; Accepted April 9, 2018; Epub May 1, 2018; Published May 15, 2018 Abstract: Glioma, the most prevalent malignancy in brain, is classified into four grades (I, II, III, and IV), and grade IV glioma is also known as glioblastoma multiforme (GBM). Aberrant activation of receptor tyrosine kinases (RTKs), including platelet-derived growth factor receptor (PDGFR), are frequently observed in glioma. Accumulating evi- dence suggests that PDGFR plays critical roles during glioma development and progression and is a promising drug target for GBM therapy. However, PDGFR inhibitor (PDGFRi) has failed in clinical trials, at least partially, due to the activation of other RTKs, which compensates for PDGFR inhibition and renders tumor cells resistance to PDGFRi. Therefore, identifying the RTKs responsible for PDGFRi resistance might provide new therapeutic targets to syner- getically enhance the efficacy of PDGFRi. -

Rat FLT4 / VEGFR3 ELISA Kit (ARG82090)
Product datasheet [email protected] ARG82090 Package: 96 wells Rat FLT4 / VEGFR3 ELISA Kit Store at: 4°C Component Cat. No. Component Name Package Temp ARG82090-001 Antibody-coated 8 X 12 strips 4°C. Unused strips microplate should be sealed tightly in the air-tight pouch. ARG82090-002 Standard 2 X 10 ng/vial 4°C ARG82090-003 Standard/Sample 30 ml (Ready to use) 4°C diluent ARG82090-004 Antibody conjugate 1 vial (100 µl) 4°C concentrate (100X) ARG82090-005 Antibody diluent 12 ml (Ready to use) 4°C buffer ARG82090-006 HRP-Streptavidin 1 vial (100 µl) 4°C concentrate (100X) ARG82090-007 HRP-Streptavidin 12 ml (Ready to use) 4°C diluent buffer ARG82090-008 25X Wash buffer 20 ml 4°C ARG82090-009 TMB substrate 10 ml (Ready to use) 4°C (Protect from light) ARG82090-010 STOP solution 10 ml (Ready to use) 4°C ARG82090-011 Plate sealer 4 strips Room temperature Summary Product Description ARG82090 Rat FLT4 / VEGFR3 ELISA Kit is an Enzyme Immunoassay kit for the quantification of Rat FLT4 / VEGFR3 in serum and cell culture supernatants. Tested Reactivity Rat Tested Application ELISA Specificity There is no detectable cross-reactivity with other relevant proteins. Target Name FLT4 / VEGFR3 Conjugation HRP Conjugation Note Substrate: TMB and read at 450 nm. Sensitivity 78 pg/ml Sample Type Serum and cell culture supernatants. Standard Range 156 - 10000 pg/ml Sample Volume 100 µl www.arigobio.com 1/3 Precision Intra-Assay CV: 5.2%; Inter-Assay CV: 6.2% Alternate Names FLT-4; FLT41; Vascular endothelial growth factor receptor 3; VEGFR3; VEGFR-3; PCL; Tyrosine-protein kinase receptor FLT4; LMPH1A; EC 2.7.10.1; Fms-like tyrosine kinase 4 Application Instructions Assay Time ~ 5 hours Properties Form 96 well Storage instruction Store the kit at 2-8°C. -

60+ Genes Tested FDA-Approved Targeted Therapies & Gene Indicators
® 60+ genes tested ABL1, ABL2, ALK, AR, ARAF, ATM, ATR, BRAF, BRCA1*, BRCA2*, BTK, CCND1, CCND2, CCND3, CDK4, Somatic mutation detection CDK6, CDKN1A, CDKN1B, CDKN2A, CDKN2B, DDR1, DDR2, EGFR, ERBB2 (HER2), ESR1, FGFR1, FGFR2, for approved cancer therapies FGFR3, FGFR4, FLCN, FLT1, FLT3, FLT4, GNA11, GNAQ, HDAC1, HDAC2, HRAS, JAK1, JAK2, KDR, KIT, in solid tumors KRAS, MAP2K1, MET, MTOR, NF1, NF2, NRAS, PALB2, PARP1, PDGFRA, PDGFRB, PIK3CA, PIK3CD, PTCH1, PTEN, RAF1, RET, ROS1, SMO, SRC, STK11, TNK2, TSC1, TSC2 FDA-approved Targeted Therapies & Gene Indicators Abiraterone AR Necitumumab EGFR Ado-Trastuzumab ERBB2 (HER2) Nilotinib ABL1, ABL2, DDR1, DDR2, KIT, PDGFRA, PDGFRB Emtansine FGFR1, FGFR2, FGFR3, FLT1, FLT4, KDR, PDGFRA, Afatinib EGFR, ERBB2 (HER2) Nintedanib PDGFRB Alectinib ALK Olaparib ATM, ATR, BRCA1*, BRCA2*, PALB2, PARP1 Anastrozole ESR1 Osimertinib EGFR Axitinib FLT1, FLT4, KDR, KIT, PDGFRA, PDGFRB CDK4, CDK6, CCND1, CCND2, CCND3, CDKN1A, Palbociclib Belinostat HDAC1, HDAC2 CDKN1B, CDKN2A, CDKN2B Panitumumab EGFR Bicalutamide AR Panobinostat HDAC1, HDAC2 Bosutinib ABL1, SRC Pazopanib FLT1, FLT4, KDR, KIT, PDGFRA, PDGFRB Cabozantinib FLT1, FLT3, FLT4, KDR, KIT, MET, RET Pertuzumab ERBB2 (HER2) Ceritinib ALK ABL1, FGFR1, FGFR2, FGFR3, FGFR4, FLT1, FLT3, FLT4, Cetuximab EGFR Ponatinib KDR, KIT, PDGFRA, PDGFRB, RET, SRC Cobimetinib MAP2K1 Ramucirumab KDR Crizotinib ALK, MET, ROS1 Regorafenib ARAF, BRAF, FLT1, FLT4, KDR, KIT, PDGFRB, RAF1, RET Dabrafenib BRAF Ruxolitinib JAK1, JAK2 Dasatinib ABL1, ABL2, DDR1, DDR2, SRC, TNK2 -

Erbb3 Is Involved in Activation of Phosphatidylinositol 3-Kinase by Epidermal Growth Factor STEPHEN P
MOLECULAR AND CELLULAR BIOLOGY, June 1994, p. 3550-3558 Vol. 14, No. 6 0270-7306/94/$04.00+0 Copyright C 1994, American Society for Microbiology ErbB3 Is Involved in Activation of Phosphatidylinositol 3-Kinase by Epidermal Growth Factor STEPHEN P. SOLTOFF,l* KERMIT L. CARRAWAY III,1 S. A. PRIGENT,2 W. G. GULLICK,2 AND LEWIS C. CANTLEY' Division of Signal Transduction, Department ofMedicine, Beth Israel Hospital, Boston, Massachusetts 02115,1 and Molecular Oncology Laboratory, ICRF Oncology Group, Hammersmith Hospital, London W12 OHS, United Kingdom2 Received 11 October 1993/Returned for modification 11 November 1993/Accepted 24 February 1994 Conflicting results concerning the ability of the epidermal growth factor (EGF) receptor to associate with and/or activate phosphatidylinositol (Ptdlns) 3-kinase have been published. Despite the ability of EGF to stimulate the production of Ptdlns 3-kinase products and to cause the appearance of PtdIns 3-kinase activity in antiphosphotyrosine immunoprecipitates in several cell lines, we did not detect EGF-stimulated Ptdlns 3-kinase activity in anti-EGF receptor immunoprecipitates. This result is consistent with the lack of a phosphorylated Tyr-X-X-Met motif, the p85 Src homology 2 (SH2) domain recognition sequence, in this receptor sequence. The EGF receptor homolog, ErbB2 protein, also lacks this motif. However, the ErbB3 protein has seven repeats of the Tyr-X-X-Met motif in the carboxy-terminal unique domain. Here we show that in A431 cells, which express both the EGF receptor and ErbB3, Ptdlns 3-kinase coprecipitates with the ErbB3 protein (pl80eR3) in response to EGF. p180B3 is also shown to be tyrosine phosphorylated in response to EGF. -

RET Gene Fusions in Malignancies of the Thyroid and Other Tissues
G C A T T A C G G C A T genes Review RET Gene Fusions in Malignancies of the Thyroid and Other Tissues Massimo Santoro 1,*, Marialuisa Moccia 1, Giorgia Federico 1 and Francesca Carlomagno 1,2 1 Department of Molecular Medicine and Medical Biotechnology, University of Naples “Federico II”, 80131 Naples, Italy; [email protected] (M.M.); [email protected] (G.F.); [email protected] (F.C.) 2 Institute of Endocrinology and Experimental Oncology of the CNR, 80131 Naples, Italy * Correspondence: [email protected] Received: 10 March 2020; Accepted: 12 April 2020; Published: 15 April 2020 Abstract: Following the identification of the BCR-ABL1 (Breakpoint Cluster Region-ABelson murine Leukemia) fusion in chronic myelogenous leukemia, gene fusions generating chimeric oncoproteins have been recognized as common genomic structural variations in human malignancies. This is, in particular, a frequent mechanism in the oncogenic conversion of protein kinases. Gene fusion was the first mechanism identified for the oncogenic activation of the receptor tyrosine kinase RET (REarranged during Transfection), initially discovered in papillary thyroid carcinoma (PTC). More recently, the advent of highly sensitive massive parallel (next generation sequencing, NGS) sequencing of tumor DNA or cell-free (cfDNA) circulating tumor DNA, allowed for the detection of RET fusions in many other solid and hematopoietic malignancies. This review summarizes the role of RET fusions in the pathogenesis of human cancer. Keywords: kinase; tyrosine kinase inhibitor; targeted therapy; thyroid cancer 1. The RET Receptor RET (REarranged during Transfection) was initially isolated as a rearranged oncoprotein upon the transfection of a human lymphoma DNA [1]. -

RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients Shumei Kato1, Vivek Subbiah2, Erica Marchlik3, Sheryl K
Published OnlineFirst September 28, 2016; DOI: 10.1158/1078-0432.CCR-16-1679 Personalized Medicine and Imaging Clinical Cancer Research RET Aberrations in Diverse Cancers: Next-Generation Sequencing of 4,871 Patients Shumei Kato1, Vivek Subbiah2, Erica Marchlik3, Sheryl K. Elkin3, Jennifer L. Carter3, and Razelle Kurzrock1 Abstract Purpose: Aberrations in genetic sequences encoding the tyrosine (52/88)], cell cycle–associated genes [39.8% (35/88)], the PI3K kinase receptor RET lead to oncogenic signaling that is targetable signaling pathway [30.7% (27/88)], MAPK effectors [22.7% with anti-RET multikinase inhibitors. Understanding the compre- (20/88)], or other tyrosine kinase families [21.6% (19/88)]. hensive genomic landscape of RET aberrations across multiple RET fusions were mutually exclusive with MAPK signaling cancers may facilitate clinical trial development targeting RET. pathway alterations. All 72 patients harboring coaberrations Experimental Design: We interrogated the molecular portfolio had distinct genomic portfolios, and most [98.6% (71/72)] of 4,871 patients with diverse malignancies for the presence of had potentially targetable coaberrations with either an FDA- RET aberrations using Clinical Laboratory Improvement Amend- approved or an investigational agent. Two cases with lung ments–certified targeted next-generation sequencing of 182 or (KIF5B-RET) and medullary thyroid carcinoma (RET M918T) 236 gene panels. thatrespondedtoavandetanib(multikinase RET inhibitor)- Results: Among diverse cancers, RET aberrations were iden- containing regimen are shown. tified in 88 cases [1.8% (88/4, 871)], with mutations being Conclusions: RET aberrations were seen in 1.8% of diverse the most common alteration [38.6% (34/88)], followed cancers, with most cases harboring actionable, albeit dis- by fusions [30.7% (27/88), including a novel SQSTM1-RET] tinct, coexisting alterations. -
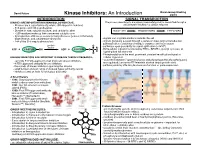
Kinase Inhibitors: an Introduction
David Peters Baran Group Meeting Kinase Inhibitors: An Introduction 2/2/19 INTRODUCTION SIGNAL TRANSDUCTION KINASES ARE IMPORTANT IN HUMAN BIOLOGY/DISEASE: The process describing how a signal (chemical/physical) is transmitted through a - Kinases are a superfamily of proteins (5th largest in humans) cell ultimately resulting in a cellular response - 518 genes and 106 pseudogenes - Diverse in size, subunit structure, and cellular location RECEPTOR TRANSDUCERS EFFECTORS - ~260 residues make up their conserved catalytic core - dysregulation of kinases occurs in many diseases (cancer, inflamatory, degenerative, and autoimmune diseases) - signals can originate inside or outside the cell - 244 of the 518 map to disease loci - signals generally passed through a series of steps (signal transduction pathway) often consisting of multiple enzymes and messengers protein O - pathways open possibility for signal aplification (>1x106) kinase ATP + PROTEIN OH ADP + PROTEIN O P O - Extracellular signals transduced by RTKs, GPCR’s, guanlyl cyclases, or ligand-gated Ion channels O - Phosphorylation is the most prominent covalent modification/signal in KINASES INHIBITORS ARE IMPORTANT IN DISEASE THERAPY/RESEARCH: cellular regulation - currently 51 FDA approved small molecule kinase inhibitors - “converter enzymes” (protein kinases and phosphoprotein phosphorlyases) - 4 FDA approved antibody kinase inhibitors are regulated; conserve ATP/maintain desired target protein state - thousands of known inhibitors spanning the kinome - pathway ends by affecting biomolecule -

A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers
cancers Article A Tyrosine Kinase Expression Signature Predicts the Post-Operative Clinical Outcome in Triple Negative Breast Cancers Alexandre de Nonneville 1, Pascal Finetti 2 , José Adelaide 2, Éric Lambaudie 3, Patrice Viens 1, Anthony Gonçalves 1 , Daniel Birnbaum 2, Emilie Mamessier 2 and François Bertucci 1,2,* 1 Department of Medical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CRCM, CNRS, INSERM, 13000 Marseille, France 2 Laboratory of Predictive Oncology, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, Inserm UMR1068, CNRS UMR725, Aix-Marseille Université, 13000 Marseille, France 3 Department of Surgical Oncology, Institut Paoli-Calmettes, Aix-Marseille Univ, CNRS, INSERM, CRCM, 13000 Marseille, France * Correspondence: [email protected]; Tel.: +33-4-91-22-35-37; Fax: +33-4-91-22-36-70 Received: 15 July 2019; Accepted: 9 August 2019; Published: 13 August 2019 Abstract: Triple negative breast cancer (TNBC) represent 15% of breast cancers. Histoclinical features and marketed prognostic gene expression signatures (GES) failed to identify good- and poor-prognosis patients. Tyrosine kinases (TK) represent potential prognostic and/or therapeutic targets for TNBC. We sought to define a prognostic TK GES in a large series of TNBC. mRNA expression and histoclinical data of 6379 early BCs were collected from 16 datasets. We searched for a TK-based GES associated with disease-free survival (DFS) and tested its robustness in an independent validation set. A total of 1226 samples were TNBC. In the learning set of samples (N = 825), we identified a 13-TK GES associated with DFS. This GES was associated with cell proliferation and immune response. -

Functional Analysis of Somatic Mutations Affecting Receptor Tyrosine Kinase Family in Metastatic Colorectal Cancer
Author Manuscript Published OnlineFirst on March 29, 2019; DOI: 10.1158/1535-7163.MCT-18-0582 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Functional analysis of somatic mutations affecting receptor tyrosine kinase family in metastatic colorectal cancer Leslie Duplaquet1, Martin Figeac2, Frédéric Leprêtre2, Charline Frandemiche3,4, Céline Villenet2, Shéhérazade Sebda2, Nasrin Sarafan-Vasseur5, Mélanie Bénozène1, Audrey Vinchent1, Gautier Goormachtigh1, Laurence Wicquart6, Nathalie Rousseau3, Ludivine Beaussire5, Stéphanie Truant7, Pierre Michel8, Jean-Christophe Sabourin9, Françoise Galateau-Sallé10, Marie-Christine Copin1,6, Gérard Zalcman11, Yvan De Launoit1, Véronique Fafeur1 and David Tulasne1 1 Univ. Lille, CNRS, Institut Pasteur de Lille, UMR 8161 - M3T – Mechanisms of Tumorigenesis and Target Therapies, F-59000 Lille, France. 2 Univ. Lille, Plateau de génomique fonctionnelle et structurale, CHU Lille, F-59000 Lille, France 3 TCBN - Tumorothèque Caen Basse-Normandie, F-14000 Caen, France. 4 Réseau Régional de Cancérologie – OncoBasseNormandie – F14000 Caen – France. 5 Normandie Univ, UNIROUEN, Inserm U1245, IRON group, Rouen University Hospital, Normandy Centre for Genomic and Personalized Medicine, F-76000 Rouen, France. 6 Tumorothèque du C2RC de Lille, F-59037 Lille, France. 7 Department of Digestive Surgery and Transplantation, CHU Lille, Univ Lille, 2 Avenue Oscar Lambret, 59037, Lille Cedex, France. 8 Department of hepato-gastroenterology, Rouen University Hospital, Normandie Univ, UNIROUEN, Inserm U1245, IRON group, F-76000 Rouen, France. 9 Department of Pathology, Normandy University, INSERM 1245, Rouen University Hospital, F 76 000 Rouen, France. 10 Department of Pathology, MESOPATH-MESOBANK, Centre León Bérard, Lyon, France. 11 Thoracic Oncology Department, CIC1425/CLIP2 Paris-Nord, Hôpital Bichat-Claude Bernard, Paris, France.