Zooming in on the Lettuce Genome: Species Relationships Inlactuca S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Resources Collections of Leafy Vegetables (Lettuce, Spinach, Chicory, Artichoke, Asparagus, Lamb's Lettuce, Rhubarb An
Genet Resour Crop Evol (2012) 59:981–997 DOI 10.1007/s10722-011-9738-x RESEARCH ARTICLE Genetic resources collections of leafy vegetables (lettuce, spinach, chicory, artichoke, asparagus, lamb’s lettuce, rhubarb and rocket salad): composition and gaps R. van Treuren • P. Coquin • U. Lohwasser Received: 11 January 2011 / Accepted: 21 July 2011 / Published online: 7 August 2011 Ó The Author(s) 2011. This article is published with open access at Springerlink.com Abstract Lettuce, spinach and chicory are gener- nl/cgn/pgr/LVintro/. Based on a literature study, an ally considered the main leafy vegetables, while a analysis of the gene pool structure of the crops was fourth group denoted by ‘minor leafy vegetables’ performed and an inventory was made of the distri- includes, amongst others, rocket salad, lamb’s lettuce, bution areas of the species involved. The results of asparagus, artichoke and rhubarb. Except in the case these surveys were related to the contents of the of lettuce, central crop databases of leafy vegetables newly established databases in order to identify the were lacking until recently. Here we report on the main collection gaps. Priorities are presented for update of the international Lactuca database and the future germplasm acquisition aimed at improving the development of three new central crop databases for coverage of the crop gene pools in ex situ collections. each of the other leafy vegetable crop groups. Requests for passport data of accessions available Keywords Chicory Á Crop database Á Germplasm to the user community were addressed to all known availability Á Lettuce Á Minor leafy vegetables Á European collection holders and to the main collec- Spinach tion holders located outside Europe. -

Nabalus Racemosus (Michx.) Hook. Glaucous White Lettuce
New England Plant Conservation Program Nabalus racemosus (Michx.) Hook. Glaucous white lettuce Conservation and Research Plan for New England Prepared by: Lisa St. Hilaire Ecologist 14 Prospect St. Augusta, Maine 04339 USA For: New England Wild Flower Society 180 Hemenway Road Framingham, Massachusetts 01701 USA 508/877-7630 e-mail: [email protected] • website: www.newfs.org Approved, Regional Advisory Council, December 2003 1 SUMMARY Nabalus racemosus (Michx.) Hook., glaucous white lettuce, is a perennial member of the Asteraceae or composite family. It is considered globally secure (G5), but in New England, it is known only from northern Maine, primarily along the St. John River. There are also several occurrences along the Aroostook River. Ice scour and flooding are common annual disturbances on these rivers. Many of the N. racemosus populations were discovered during survey efforts for Pedicularis furbishiae (Furbish’s lousewort), and both species, as well as many other rarities, may be found at some sites. In other parts of the country, N. racemosus grows in prairie communities. There are currently 31 extant occurrences in Maine, 28 of these along the St. John River, and three on the Aroostook River. There are four historic occurrences, all on the Aroostook River, and one extirpated population on the Aroostook River. Nabalus racemosus is a species of Special Concern in Maine, where it is ranked S3. Other nearby areas from which it is recorded include New Brunswick (S3), Nova Scotia (S1), Newfoundland Island (S1S2), Labrador (SR), Quebec (SR), Vermont (SR), New Jersey (SH), New York (SX), and Pennsylvania (SX). Little is known regarding the biology of Nabalus racemosus. -

Gathered Food Plants in the Mountains
34063_Rivera.qxd 7/2/07 2:03 PM Page 1 Gathered Food Plants in the Mountains of Castilla–La Mancha (Spain): Ethnobotany and Multivariate Analysis1 Diego Rivera*,2, Concepción Obón3, Cristina Inocencio3, Michael Heinrich4, Alonso Verde2, José Fajardo2, and José Antonio Palazón5 2 Departamento de Biología Vegetal, Facultad de Biología, Universidad de Murcia, 30100 Murcia, Spain 3 Departamento de Biología Aplicada, EPSO, Universidad Miguel Hernández, 03312 Orihuela, Alicante, Spain 4 Centre for Pharmacognosy and Phytotherapy, The School of Pharmacy, Univ. London, 29–39 Brunswick Sq. London, WC1N 1AX, United Kingdom 5 Departamento de Ecología e Hidrología, Facultad de Biología, Universidad de Murcia, 30100 Murcia, Spain * Corresponding author: Departamento de Biología Vegetal, Facultad de Biología, Universidad de Murcia, 30100 Murcia, Spain; e-mail: [email protected] GATHERED FOOD PLANTS IN THE MOUNTAINS OF CASTILLA–LA MANCHA (SPAIN): ETHNOBOTANY AND MULTIVARIATE ANALYSIS. Gathered food plants (GFPs) (wild and weeds) are crucial for under- standing traditional Mediterranean diets. Combining open interviews and free–listing ques- tionnaires, we identified 215 GFP items, i.e., 53 fungi and 162 from 154 vascular plant species. The variation in frequency and in salience among the items follows a rectangular hyperbola. Highly salient species were Silene vulgaris (Moench) Garcke, Scolymus hispani- cus L., and Pleurotus eryngii (DC.: Fr.) Quélet. Salience and frequency showed no correlation with the expected health benefits of each species. Regional frequency in the Mediter- ranean and local frequency are directly related. Thus, local food plants are much less “local” than expected. Different types of culinary preparations provide the most information in the cluster analysis of variables. -

The Vascular Flora of Rarău Massif (Eastern Carpathians, Romania). Note Ii
Memoirs of the Scientific Sections of the Romanian Academy Tome XXXVI, 2013 BIOLOGY THE VASCULAR FLORA OF RARĂU MASSIF (EASTERN CARPATHIANS, ROMANIA). NOTE II ADRIAN OPREA1 and CULIŢĂ SÎRBU2 1 “Anastasie Fătu” Botanical Garden, Str. Dumbrava Roşie, nr. 7-9, 700522–Iaşi, Romania 2 University of Agricultural Sciences and Veterinary Medicine Iaşi, Faculty of Agriculture, Str. Mihail Sadoveanu, nr. 3, 700490–Iaşi, Romania Corresponding author: [email protected] This second part of the paper about the vascular flora of Rarău Massif listed approximately half of the whole number of the species registered by the authors in their field trips or already included in literature on the same area. Other taxa have been added to the initial list of plants, so that, the total number of taxa registered by the authors in Rarău Massif amount to 1443 taxa (1133 species and 310 subspecies, varieties and forms). There was signaled out the alien taxa on the surveyed area (18 species) and those dubious presence of some taxa for the same area (17 species). Also, there were listed all the vascular plants, protected by various laws or regulations, both internal or international, existing in Rarău (i.e. 189 taxa). Finally, there has been assessed the degree of wild flora conservation, using several indicators introduced in literature by Nowak, as they are: conservation indicator (C), threat conservation indicator) (CK), sozophytisation indicator (W), and conservation effectiveness indicator (E). Key words: Vascular flora, Rarău Massif, Romania, conservation indicators. 1. INTRODUCTION A comprehensive analysis of Rarău flora, in terms of plant diversity, taxonomic structure, biological, ecological and phytogeographic characteristics, as well as in terms of the richness in endemics, relict or threatened plant species was published in our previous note (see Oprea & Sîrbu 2012). -

Phylogenetic Relationships Within Lactuca L. (Asteraceae), Including African Species, Based on Chloroplast DNA Sequence Comparisons
Genet Resour Crop Evol DOI 10.1007/s10722-015-0332-5 RESEARCH ARTICLE Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons Zhen Wei . Shi-Xin Zhu . R. G. Van den Berg . Freek T. Bakker . M. Eric Schranz Received: 25 April 2015 / Accepted: 29 September 2015 Ó The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Lettuce (Lactuca sativa L.) belongs to the maximum likelihood and Bayesian inference analyses. genus Lactuca L. and is an important vegetable world- Biogeographical, chromosomal and morphological wide. Over the past decades, there have been many character states were reconstructed over the Bayesian controversies about the phylogeny of Lactuca species tree topology. We conclude that Lactuca contains two due to their complex and diverse morphological distinct phylogenetic clades—the crop clade and the characters and insufficient molecular sampling. In this Pterocypsela clade. Other North American, Asian and study we provide the most extensive molecular widespread species either form smaller clades or mix phylogenetic reconstruction of Lactuca, including with the Melanoseris species. The newly sampled African wild species, using two chloroplast genes African endemic species probably should be treated as (ndhF and trnL-F). The sampling covers nearly 40 % a new genus. of the total endemic African Lactuca species and 34 % of the total Lactuca species. DNA sequences from all Keywords African Lactuca Á Lactuca phylogeny Á the subfamilies of Asteraceae in Genebank and those Lettuce Á ndhF Á Phylogenetic relationships Á trnL-F generated from Lactuca herbarium samples were used to establish the affiliation of Lactuca within Astera- caeae. -

Central European Vegetation
Plant Formations in the Central European BioProvince Peter Martin Rhind Central European Beech Woodlands Beech (Fagus sylvatica) woods form the natural climax over much of Central Europe where the soils are relatively dry and can extend well into the uplands in the more southern zones. In the north, however, around Sweden it is confined to the lowlands. Beech woodlands are often open with a poorly developed shrub layer, Characteristic ground layer species may include various helleborines such as Cephalanthera damasonium, C. longifolia and C. rubra and sedges such as Carex alba, whilst in others, grasses like Sesleria caerlea or Melica uniflora may predominate, but in some of the more acidic examples, Luzula luzuloides is likely to dominate. There are also a number of endemic ground layer species. For example, in Carpathian beech woods endemics such as Dentaria glandulosa (Brassicaceae), Symphytum cordata (Boraginaceae) and the fern Polystichum braunii (Dryopteridaceae) may be encountered. Fine examples of primeaval beech woods can be found in the limestone Alps of lower Austria including the famous ‘Rothwald’ on the southeastern slopes of Dürrentein near Lunz. These range in altitude from about 940-1480 m. Here the canopy is dominated by Fagus sylvatica together with Acer pseudoplatanus, Picea abies, Ulmus glabra, and on the more acidic soils by Abies alba. Typical shrubs include Daphne mezereum, Lonicera alpigena and Rubus hirtus. At ground level the herb layer is very rich supporting possibly up to a 100 species of vascular plants. Examples include Adenostyles alliariae, Asplenium viridis, Campanula scheuchzeri, Cardamine trifolia, Cicerbita alpina, Denteria enneaphyllos, Euphorbia amygdaloides, Galium austriacum, Homogyne alpina, Lycopodium annotinum, Mycelis muralis, Paris quadrifolia, Phyteuma spicata, Prenanthes purpurea, Senecio fuchsii, Valeriana tripteris, Veratrum album and the central European endemic Helliborus niger (Ranunculaceae). -

Genetic Diversity and Evolution in Lactuca L. (Asteraceae)
Genetic diversity and evolution in Lactuca L. (Asteraceae) from phylogeny to molecular breeding Zhen Wei Thesis committee Promotor Prof. Dr M.E. Schranz Professor of Biosystematics Wageningen University Other members Prof. Dr P.C. Struik, Wageningen University Dr N. Kilian, Free University of Berlin, Germany Dr R. van Treuren, Wageningen University Dr M.J.W. Jeuken, Wageningen University This research was conducted under the auspices of the Graduate School of Experimental Plant Sciences. Genetic diversity and evolution in Lactuca L. (Asteraceae) from phylogeny to molecular breeding Zhen Wei Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr A.P.J. Mol, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Monday 25 January 2016 at 1.30 p.m. in the Aula. Zhen Wei Genetic diversity and evolution in Lactuca L. (Asteraceae) - from phylogeny to molecular breeding, 210 pages. PhD thesis, Wageningen University, Wageningen, NL (2016) With references, with summary in Dutch and English ISBN 978-94-6257-614-8 Contents Chapter 1 General introduction 7 Chapter 2 Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons* 31 Chapter 3 Phylogenetic analysis of Lactuca L. and closely related genera (Asteraceae), using complete chloroplast genomes and nuclear rDNA sequences 99 Chapter 4 A mixed model QTL analysis for salt tolerance in -

5. Tribe CICHORIEAE 菊苣族 Ju Ju Zu Shi Zhu (石铸 Shih Chu), Ge Xuejun (葛学军); Norbert Kilian, Jan Kirschner, Jan Štěpánek, Alexander P
Published online on 25 October 2011. Shi, Z., Ge, X. J., Kilian, N., Kirschner, J., Štěpánek, J., Sukhorukov, A. P., Mavrodiev, E. V. & Gottschlich, G. 2011. Cichorieae. Pp. 195–353 in: Wu, Z. Y., Raven, P. H. & Hong, D. Y., eds., Flora of China Volume 20–21 (Asteraceae). Science Press (Beijing) & Missouri Botanical Garden Press (St. Louis). 5. Tribe CICHORIEAE 菊苣族 ju ju zu Shi Zhu (石铸 Shih Chu), Ge Xuejun (葛学军); Norbert Kilian, Jan Kirschner, Jan Štěpánek, Alexander P. Sukhorukov, Evgeny V. Mavrodiev, Günter Gottschlich Annual to perennial, acaulescent, scapose, or caulescent herbs, more rarely subshrubs, exceptionally scandent vines, latex present. Leaves alternate, frequently rosulate. Capitulum solitary or capitula loosely to more densely aggregated, sometimes forming a secondary capitulum, ligulate, homogamous, with 3–5 to ca. 300 but mostly with a few dozen bisexual florets. Receptacle naked, or more rarely with scales or bristles. Involucre cylindric to campanulate, ± differentiated into a few imbricate outer series of phyllaries and a longer inner series, rarely uniseriate. Florets with 5-toothed ligule, pale yellow to deep orange-yellow, or of some shade of blue, including whitish or purple, rarely white; anthers basally calcarate and caudate, apical appendage elongate, smooth, filaments smooth; style slender, with long, slender branches, sweeping hairs on shaft and branches; pollen echinolophate or echinate. Achene cylindric, or fusiform to slenderly obconoidal, usually ribbed, sometimes compressed or flattened, apically truncate, attenuate, cuspi- date, or beaked, often sculptured, mostly glabrous, sometimes papillose or hairy, rarely villous, sometimes heteromorphic; pappus of scabrid [to barbellate] or plumose bristles, rarely of scales or absent. -

Additions to the New Flora of Vermont
Gilman, A.V. Additions to the New Flora of Vermont. Phytoneuron 2016-19: 1–16. Published 3 March 2016. ISSN 2153 733X ADDITIONS TO THE NEW FLORA OF VERMONT ARTHUR V. GILMAN Gilman & Briggs Environmental 1 Conti Circle, Suite 5, Barre, Vermont 05641 [email protected] ABSTRACT Twenty-two species of vascular plants are reported for the state of Vermont, additional to those reported in the recently published New Flora of Vermont. These are Agrimonia parviflora, Althaea officinalis , Aralia elata , Beckmannia syzigachne , Bidens polylepis , Botrychium spathulatum, Carex panicea , Carex rostrata, Eutrochium fistulosum , Ficaria verna, Hypopitys lanuginosa, Juncus conglomeratus, Juncus diffusissimus, Linum striatum, Lipandra polysperma , Matricaria chamomilla, Nabalus racemosus, Pachysandra terminalis, Parthenocissus tricuspidata , Ranunculus auricomus , Rosa arkansana , and Rudbeckia sullivantii. Also new are three varieties: Crataegus irrasa var. irrasa , Crataegus pruinosa var. parvula , and Viola sagittata var. sagittata . Three species that have been reported elsewhere in 2013–2015, Isoetes viridimontana, Naias canadensis , and Solidago brendiae , are also recapitulated. This report and the recently published New Flora of Vermont (Gilman 2015) together summarize knowledge of the vascular flora of Vermont as of this date. The New Flora of Vermont was recently published by The New York Botanical Garden Press (Gilman 2015). It is the first complete accounting of the vascular flora of Vermont since 1969 (Seymour 1969) and adds more than 200 taxa to the then-known flora of the state. However, the manuscript for the New Flora was finalized in spring 2013 and additional species are now known: those that have been observed more recently, that have been recently encountered (or re-discovered) in herbaria, or that were not included because they were under study at the time of finalization. -

Cicerbita Alpina) 5
FUTURBIOERBE 2010-2013 1 FuturBioErbe Futur: the future of the agriculture Bio: organic farming crop Erbe: the farming of wild plants related to tradition 2 Project promoted by Friuli Venezia Giulia Region L.R. n. 26/05 art. 17 3 Partners: Istituto IPSSA Pozzuolo 4 THE PURPOSE OF THE PROJECTS PICKING UP OF WILD PLANTS FARMING TRADITION INNOVACTION 5 EXPERIMENTAL STEPS RESEARCH FARMS VIVAISTIC TECNIQUES PRODUCT AGRONOMIC MODELS 6 PROJECT STEPS Creation of a productive pathway Farmer Restaurateur Hobbyist Transformer Picker Added value: Market •Quality •Traceability •Guarantee •Synergy approach 7 OUTPUTS of PROJECT Environmental Economic Technic-scientific • Recovering of marginal, • Creation of an • Determination of the not farmed or discarded additional income for best agronomic coltivation fields farmers techniques • Preservation of the • Increasing and • Creation of biodiversity consolidation of a experimental field to test • Dissemination of some “niche” market different low impact regional vegetal • Characterisation of agronomic techniques varieties with high low valorized products • Evaluation of the effects productive potenziality • Development of a of the picking up of highly • New synergies substainable agriculture extintion risk between agriculture and to create high quality plantsduzione territory products with a high • Definition of market market value startegies 8 PATHWAY of THE TRIAL Integration and promotion of the touristic offert Tight juncture between agriculture and eno-gastronomic offert Consolidation of the agricultural -
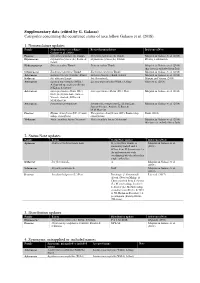
Categories Concerning the Occurrence Status of Taxa Follow Galasso Et Al
Supplementary data (edited by G. Galasso) Categories concerning the occurrence status of taxa follow Galasso et al. (2018). 1. Nomenclature updates Family Nomenclature according to Revised nomenclature References/Note Galasso et al. (2018) Poaceae Cenchrus purpurascens Thumb. Cenchrus purpurascens Thunb. Misprint in Galasso et al. (2018) Dipsacaceae Cephalaria syriaca (L.) Roem. & Cephalaria syriaca (L.) Schrad. Priority combination Schult. Hydrangeaceae Deutzia scabra Thumb. Deutzia scabra Thunb. Misprint in Galasso et al. (2018); this taxon is excluded from Italy Celastraceae Euonymus japonicus Thumb. Euonymus japonicus Thunb. Misprint in Galasso et al. (2018) Asteraceae Gazania linearis (Thumb.) Druce Gazania linearis (Thunb.) Druce Misprint in Galasso et al. (2018) Iridaceae Iris albicans Lange Iris florentina L. Martini and Viciani (2018) Asteraceae Lactuca macrophylla (Willd.) Lactuca macrophylla (Willd.) A.Gray Güzel et al. (2018) A.Gray subsp. uralensis (Rouy) N.Kilian & Greuter Asteraceae Osteospermum ecklonis (DC.) Osteospermum ecklonis (DC.) Norl. Misprint in Galasso et al. (2018) Norl. (L.) D.Gut.Larr., Santos- Vicente, Anderb., E.Rico & M.M.Mart.Ort. Asteraceae Pentanema germanicum Pentanema germanicum (L.) D.Gut.Larr., Misprint in Galasso et al. (2018) Santos-Vicente, Anderb., E.Rico & M.M.Mart.Ort. Poaceae Elymus obtusiflorus (DC.) Conert Thinopyrum obtusiflorum (DC.) Banfi subsp. Banfi (2018) subsp. obtusiflorus obtusiflorum Violaceae Viola cucullata Aiton (Vitaceae) Viola cucullata Aiton (Violaceae) Misprint in Galasso et al. (2018); this taxon is excluded from Italy 2. Status/Note updates Family Taxon Status/Note update References/Note Apiaceae Hydrocotyle bonariensis Lam. H. verticillata Thunb. is Misprint in Galasso et al. commonly traded, and it (2018) differs from H. bonariensis for the inflorescences with overlapping whorls rather than single umbrellas. -

Plant Communities with Naturalized Elaeagnus Angustifolia L. As a New
Acta Biologica Sibirica 7: 49–61 (2021) doi: 10.3897/abs.7.e58204 https://abs.pensoft.net RESEARCH ARTICLE Plant communities with naturalized Elaeagnus angustifolia L. as a new vegetation element in Altai Krai (Southwestern Siberia, Russia) Alena A. Shibanova1, Natalya V. Ovcharova1 1 Altai State University, 61 Lenina Prospect, Barnaul, 656049, Russia Corresponding author: Alena A. Shibanova ([email protected]) Academic editor: D. German | Received 1 September 2020 | Accepted 29 January 2021 | Published 13 April 2021 http://zoobank.org/1B70B70D-7F9C-4CEB-907A-7FA39D1446A1 Citation: Shibanova AA, Ovcharova NV (2021) Plant communities with naturalized Elaeagnus angustifolia L. as a new vegetation element in Altai Krai (Southwestern Siberia, Russia). Acta Biologica Sibirica 7: 49–61 https://doi. org/10.3897/abs.7.e58204 Abstract Elaeagnus angustifolia L. (Russian olive) is a deciduous small tree or large multi-stemmed shrub that becomes invader in different countries all other the world. It is potentially invasive in some regions of Russia. In the beginning of 20th century, it was introduced to the steppe region of Altai Krai (Rus- sia, southwestern Siberia) to prevent wind erosion. During last 20 years, Russian olive starts to create its own natural stands and to influence on native vegetation. This article presents the results of eco- coenotic survey of natural plant communities dominated by Elaeagnus angustifolia L. first described for Siberia and the analysis of their possible syntaxonomic position. The investigation conducted during summer season 2012 in the steppe region of Altai Krai allows revealing one new for Siberia association Elytrigio repentis–Elaeagnetum angustifoliae and no-ranged community Bromopsis inermis–Elae- agnus angustifolia which were included to the Class Nerio–Tamaricetea, to the Order Tamaricetalia ramosissimae.